Volume 17, Number 8—August 2011
Dispatch
Cowpox Virus in Llama, Italy
Figure 2
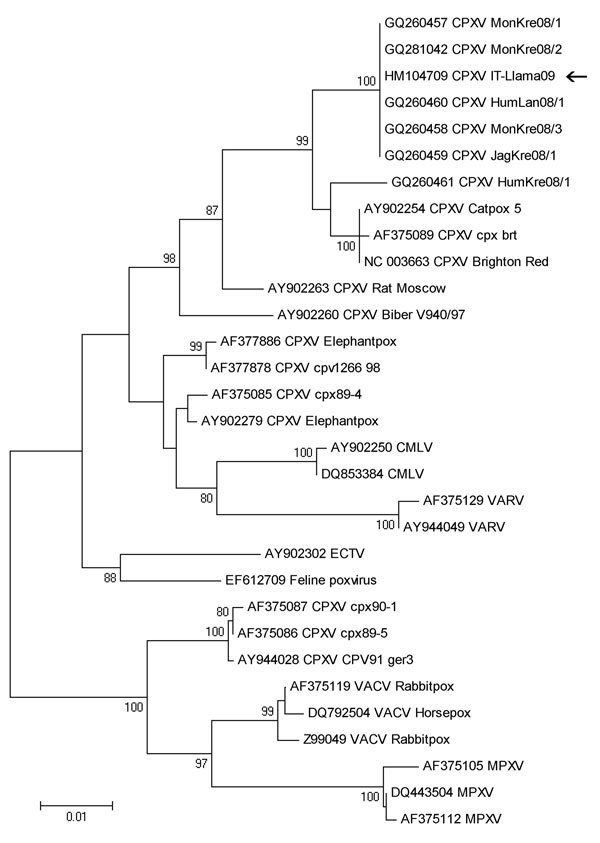
Figure 2. Phylogenetic tree based on nucleotide sequences of the complete hemagglutinin open reading frame (921 bp) from the llama orthopoxvirus isolate (arrow) and additional orthopoxvirus sequences available in GenBank. The tree has been constructed by using nucleotide alignment, the Kimura 2-parameter algorithm, and the neighbor-joining method implemented in MEGA4.1 software (www.megasoftware.net). Bootstrap values >75 are shown at nodes. CPXV, cowpox virus; CMLV, camelpox virus; VARV, variola virus; ECTV, ectromelia virus; VACV, vaccinia virus; MPXV, monkeypox virus. Scale bar indicates genetic diversity at the nucleotide level.
Page created: August 15, 2011
Page updated: August 15, 2011
Page reviewed: August 15, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.