Volume 17, Number 8—August 2011
Dispatch
Novel Lyssavirus in Natterer’s Bat, Germany
Figure 2
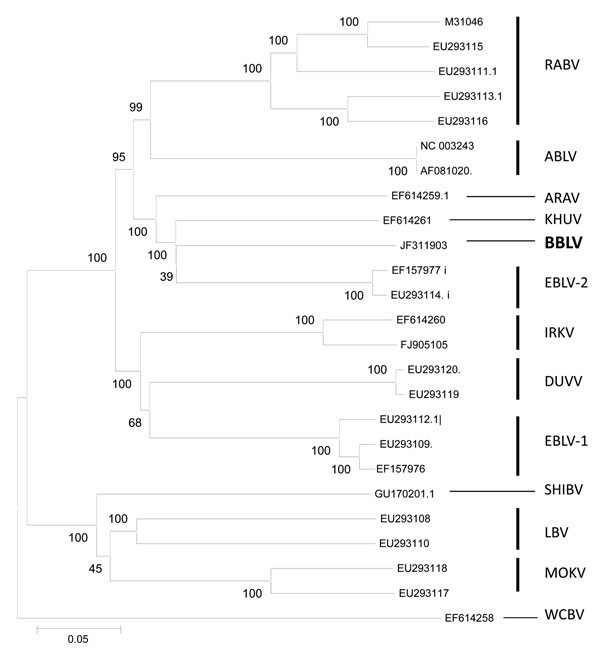
Figure 2. Phylogenetic tree inferred from concatenated N-P-M-G-L sequences of bat lyssaviruses. The neighbor-joining method (Kimura 2-parameter) was used as implemented in MEGA4 software (www.megasoftware.net). Bootstrap values (500 replicates) are shown next to branches. Scale bar indicates nucleotide substitutions per site. Virus isolated in this study is shown in boldface. RABV, rabies virus; ABLV, Australian bat lyssavirus; ARAV, Aravan virus; KHUV, Khujand virus; BBLV, Bokeloh bat lyssavirus; European bat lyssavirus; IRKV, Irkut virus; DUVV, Duvenhage virus; SHIBV, Shimoni bat virus; LBV, Lagos bat virus; MOKV, Mokola virus; WCBV, West Caucasian bat virus.
Page created: August 16, 2011
Page updated: August 16, 2011
Page reviewed: August 16, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.