Volume 20, Number 4—April 2014
Dispatch
Invasive Salmonella enterica Serotype Typhimurium Infections, Democratic Republic of the Congo, 2007–2011
Figure 2
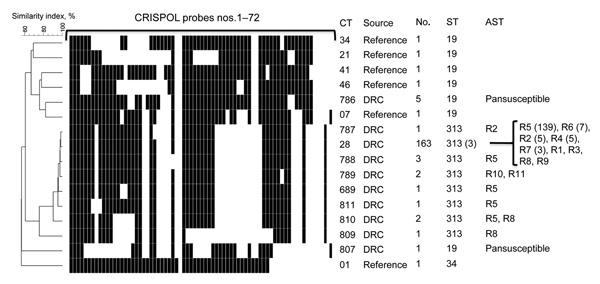
Figure 2. . Representative CRISPOL profiles of Salmonella enterica serotype Typhimurium isolates studied. CRISPOL is a recently developed high-throughput assay based on clustered regularly interspaced short palindromic repeats (CRISPR) polymorphisms. Black squares indicate presence of the CRISPR spacer, detected by the corresponding probe; white squares indicates absence of the spacer. The dendrogram was generated by using BioNumerics version 6.6 software (Applied Maths, Sint-Martens-Latem, Belgium) as described (5). The CRISPOL types (CTs) detected among the 180 isolates from the Democratic Republic of Congo (DRC) are labeled as DRC in the Source column. Six common CTs of the Pasteur Institute CRISPOL database (labeled as reference) are also shown. These CTs are from strains of serotype Typhimurium 02–1800 (CT34, DT120), 02–5270 (CT21, DT104), LT2 (CT41, DT4), 02–2561 (CT46, DT12), 02–1749 (CT7, DT14) or its monophasic variant of antigenic formula 1,4,[5],12:i:-, 07–1777 (CT1, DT193). For each distinct CT, the numbers of corresponding isolates, their sequence types (STs), and their antimicrobial drug susceptibility testing (AST) data are indicated. For the ST and AST columns, the numbers in parentheses refer to the number (>2) of tested isolates with such result. AST data are shown only for DRC isolates. The resistance types were as follows: R1, ASKTNGSulTmpC; R2, ASKTNGSulTmpCTe; R3, AC; R4, ASSulTmp; R5, ASSulTmpC; R6, ASSulTmpCNal; R7, ASSulTmpCTe; R8, ASulTmpC; R9, SSulTmpC; R10, ACroCazSKTNGSulTmpCTeAzi; and R11, ACroSKTNGSulTmpCTeNaAzi. Abbreviations used in the descriptions of resistance types are as follows: A, amoxicillin; Cro, ceftriaxone; Caz; ceftazidime; S, streptomycin; K, kanamycin; T, tobramycin; N, netilmicin; G, gentamicin; Sul, sulfamethoxazole; Tmp, trimethoprim; C, chloramphenicol; Te, tetracycline; Nal, nalidixic acid; Azi, azithromycin.
References
- Kingsley RA, Msefula CL, Thomson NR, Kariuki S, Holt KE, Gordon MA, Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 2009;19:2279–87. DOIPubMedGoogle Scholar
- Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. Invasive non-typhoidal Salmonella disease: an emerging and neglected tropical disease in Africa. Lancet. 2012;379:2489–99. DOIPubMedGoogle Scholar
- Okoro CK, Kingsley RA, Connor TR, Harris SR, Parry CM, Al-Mashhadani MN, Intra-continental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat Genet. 2012;44:1215–21. DOIPubMedGoogle Scholar
- Lunguya O, Lejon V, Phoba MF, Bertrand S, Vanhoof R, Glupczynski Y, Antimicrobial resistance in invasive non-typhoid Salmonella from the Democratic Republic of the Congo: emergence of decreased fluoroquinolone susceptibility and extended-spectrum beta lactamases. PLoS Negl Trop Dis. 2013;7:e2103. DOIPubMedGoogle Scholar
- Fabre L, Zhang J, Guigon G, Le Hello S, Guibert V, Accou-Demartin M, CRISPR typing and subtyping for improved laboratory surveillance of Salmonella infections. PLoS ONE. 2012;7:e36995. DOIPubMedGoogle Scholar
- Achtman M, Wain J, Weill FX, Nair S, Zhou Z, Sangal V, Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 2012;8:e1002776 . DOIPubMedGoogle Scholar
- Le Hello S, Harrois D, Bouchrif B, Sontag L, Elhani D, Guibert V, Highly drug-resistant Salmonella enterica serotype Kentucky ST198–X1: a microbiological study. Lancet Infect Dis. 2013;13:672–9. DOIPubMedGoogle Scholar
- Jarlier V, Nicolas MH, Fournier G, Philippon A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10:867–78. DOIPubMedGoogle Scholar
- Msefula CL, Kingsley RA, Gordon MA, Molyneux E, Molyneux ME, MacLennan CA, Genotypic homogeneity of multidrug resistant S. Typhimurium infecting distinct adult and childhood susceptibility groups in Blantyre, Malawi. PLoS ONE. 2012;7:e42085. DOIPubMedGoogle Scholar
- Wain J, Keddy KH, Hendriksen RS, Rubino S. Using next generation sequencing to tackle non-typhoidal Salmonella infections. J Infect Dev Ctries. 2013;7:1–5 .DOIPubMedGoogle Scholar
- Kariuki S, Revathi G, Kariuki N, Kiiru J, Mwituria J, Muyodi J, Invasive multidrug-resistant non-typhoidal Salmonella infections in Africa: zoonotic or anthroponotic transmission? J Med Microbiol. 2006;55:585–91 . DOIPubMedGoogle Scholar
- Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–52. DOIPubMedGoogle Scholar
- Holt KE, Parkhill J, Mazzoni CJ, Roumagnac P, Weill FX, Goodhead I, High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat Genet. 2008;40:987–93. DOIPubMedGoogle Scholar