Volume 20, Number 6—June 2014
Dispatch
Novel Human Bufavirus Genotype 3 in Children with Severe Diarrhea, Bhutan
Figure
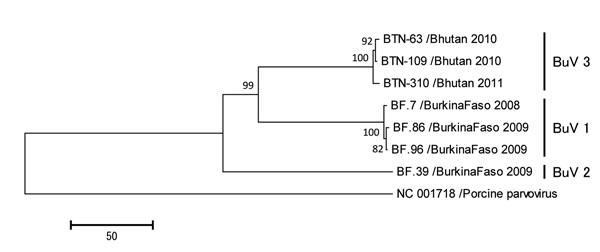
Figure. Phylogenetic trees of the viral protein 1 (VP1) of bufaviruses, constructed by using deduced amino acid sequences by neighbor-joining methodThe full length open reading frames of VP1 genes were used to deduce amino acid sequencesPorcine parvovirus strain NC 001718 was used as an out-groupThe numbers adjacent to the nodes represent the bootstrap values, and values <50% are not shownScale bar indicates the genetic distances expressed as amino acid substitutions per siteAccession numbers for DNA Data Bank of Japan, European Molecular Biology Laboratory, and GenBank are AB847987, AB847988, and AB847989 for strains BTN-63, BTN-109, and BTN-310, respectively.
Page created: May 16, 2014
Page updated: May 16, 2014
Page reviewed: May 16, 2014
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.