Volume 20, Number 6—June 2014
Letter
Rapid Metagenomic Diagnostics for Suspected Outbreak of Severe Pneumonia
Figure
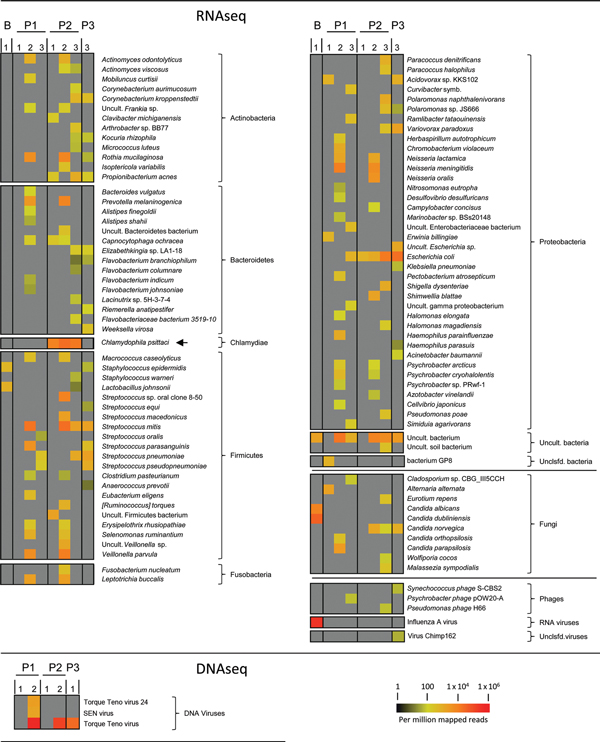
Figure. Next-generation sequencing of RNA (RNaseq) and DNA (DNaseq) isolated from bronchoalveolar lavage (BAL) samples from 3 patients with severe pneumonia, northern GermanyShown are data from BLASTN (http://blast.ncbi.nlm.nih.gov/Blast.cgi) analysis of de novo assembled sequence contigs (www.virus-genomics.org/supplementals/EID1406.pdf)Relative abundance of contig reads mapping to bacterial, fungal, or viral species is indicated by a heat map (scale bar)Gray indicates that no reads were detectedDiagnostic samples were obtained from 3 patients (lanes P1, P2, and P3)Lane B, control BAL sample (analyzed by using RNaseq only) from an influenza patient; lane1, MS: analysis on the Illumina MiSeq platform (www.illumina.com/systems.ilmn); lane 2, HS: analysis on the Illumina HiSeq platform (www.illumina.com/systems.ilmn); lane 3, HS dpl., RNA samples depleted of human rRNA before analysis on a HiSeq instrumentChlamydophila psittaci, which was unequivocally detected in all samples from patient 2 but not in samples from the other patients, is indicated by an arrowSymb., symbiont; Uncult., uncultured; Unclsfd., unclassified; SEN virus, strain of Torque teno virus.
1These authors contributed equally to this article.