Volume 22, Number 6—June 2016
Letter
Loss of 89K Pathogenicity Island in Epidemic Streptococcus suis, China
Cite This Article
Citation for Media
To the Editor: Streptococcus suis serotype 2 (SS2) is a previously neglected, newly emerging human pathogen that causes occupational and opportunistic infections (1,2). Outbreaks of fatal human SS2 infections in China, featuring streptococcal toxic shock syndrome, in 2005 seriously challenged global public health (3–5). The epidemic strain is unusual in that it contains a unique 89-kb (89K) pathogenicity island (PAI) (3,6,7). We observed the loss of genes from the 89K PAI in sporadic cases in southern China in 2007, implying the dynamic evolution of this PAI (8). Therefore, 89K PAI might be able to be used to monitor prevalent strains of S. suis in China (8).
We report 10 recurrent cases of human S. suis infections during 2008–2015 in southern China. Most of the hospitalized patients were male workers in close contact with pigs, pork products, or both. These patients typically exhibited clinical syndromes of meningitis, including headache, coma, vomiting, and fever. The bacterial strains acquired from humans were as follows: 2 isolates in 2008 (Stre08001 and Stre08002), 2 in 2009 (Stre09001 and Stre09002), 2 in 2011 (Stre11001 and Stre11002), 3 in 2013 (Stre13002, Stre13003, and Stre13004), and 1 in 2015 (Stre15001). Microbial and molecular assays proved that these clinical isolates were S. suis (Technical Appendix Figures 1, 2). Multiplex PCR–based molecular determination (16S rDNA, mrp, epf, and cps-2j) suggested that all strains except Stre13002 were SS2 (Technical Appendix Figure 2) (3). To determine whether these clinical isolates derived from the same Chinese epidemic clone 05ZYH33, we sequenced an array of virulence factor–encoding genes, as well as the 16S rDNA genes. Phylogenetic trees indicated that all 10 clinical strains are classified into the same subclade as that of the strain 05ZYH33 (Technical Appendix Figure 2).
Subsequent analyses by pulsed-field gel electrophoresis revealed that genotypes are diverse among these clinical strains, which can be roughly divided into 6 groups (Technical Appendix Figure 3). Given that the Sao surface antigen protein possesses 3 allelic variants (Sao-L [670 aa], Sao-M [580 aa], and Sao-S [489/490 aa]) (9), we thus assayed it with these clinical strains. Unexpectedly, we found 2 more new allelic variants, referred to as Sao-L1 (640 aa) and Sao-L2 (611 aa). Except for the strain BM407, which is a Chinese epidemic SS2 encoding Sao-L1, a version 30 residues shorter than Sao-L, 8 of the 10 clinical S. suis isolates consistently had the same new form of Sao protein, Sao-L2 (611 aa) (Technical Appendix Figure 4).
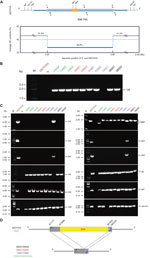
Because 89K PAI is in dynamic evolution, determining whether it remains present in clinical strains is of interest. As previously designed (Technical Appendix Table 1), a specific pair of boundary primers (1/6) was applied for PCR-based detection of the 89K PAI (Figure, panel A). In principle, the PCR-positive result suggests the absence of 89K PAI, whereas the PCR-negative result indicates the presence of 89K PAI (6,8). Unlike the epidemic strain 05ZYH33 that has the 89K PAI, 9 of the 10 clinical strains examined (Stre08001, Stre08002, Stre09001, Stre09002, Stre11001, Stre11002, Stre13002, Stre13003, and Stre13004) were unexpectedly found to be PCR positive for the unique 1/6 DNA fragment with expected size of ≈1.5 kb (Figure, panel B). This finding indicates that the 89K PAI is lost in these 9 clinical strains. We saw similar scenarios in the subsequent PCRs for other inner genes/DNA fragments (943 and 944 [10]; 1/2, 3/4, and 5/6 [8]) inside of 89K PAI (Figure, panel C). Further DNA sequencing of the 1/6 PCR product showed that it matches well with the 2 boundary regions neighboring the 89K PAI, validating the loss of 89K PAI in these 9 clinical isolates (Figure, panel D). In contrast, the strain Stre15001 behaved similarly to that of the 05ZYH33 containing the 89K PAI, in that both are PCR positive for the 4 amplicons of 1/2, 5/6, 943, and 944 but PCR negative for the 1/6 amplicon (Figure, panels B–D). The only minor difference between strains Stre15001 and 05ZYH33 lay in the 3/4 amplicon (Figure, panel C). Clearly the 3/4 DNA fragment is present in the 89K PAI from strain 05ZYH33 but not in the counterpart of the strain Stre15001 (Figure, panel C); that is, strain Stre15001 carries a variant of 89K PAI lacking (at least part of, if not all) the 3/4 DNA fragment. In terms of 89K PAI (and pulsed-field gel electrophoresis/Sao protein), we propose that a heterogeneous SS2 population is circulating in China. Also, we observe that the differentiation of bacterial virulence is related to the clinical strains using the infection model of Balb/c mice (Technical Appendix Figure 5).
In summary, the loss of 89K PAI might highlight the emergence of an epidemic SS2 population. This population appears to have genetic heterogeneity that is undergoing evolution in an adaption to some selection pressure from the environment, host restriction, or both.
Acknowledgments
This work was supported by Zhejiang Provincial Natural Science Foundation for Distinguished Young Scholars (grant no. LR15H190001), the National Natural Science Foundation of China (grant no. 31570027), and the start-up package from Zhejiang University (Y.F.). Dr. Feng is a recipient of the “Young 1000 Talents” Award.
Y.F. and Q.H. designed this project; X.S., H.Y., J.W., and Z.L. performed experiments and analyzed the data; B.C. and R.W. contributed reagents and tools; Y.F. wrote the article.
References
- Feng Y, Zhang H, Ma Y, Gao GF. Uncovering newly emerging variants of Streptococcus suis, an important zoonotic agent. Trends Microbiol. 2010;18:124–31 . DOIPubMedGoogle Scholar
- Gottschalk M, Segura M, Xu J. Streptococcus suis infections in humans: the Chinese experience and the situation in North America. Anim Health Res Rev. 2007;8:29–45. DOIPubMedGoogle Scholar
- Tang J, Wang C, Feng Y, Yang W, Song H, Chen Z, Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. [Erratum in: PLoS Med. 2006;3:e377. ]. PLoS Med. 2006;3:e151. DOIPubMedGoogle Scholar
- Ye C, Zhu X, Jing H, Du H, Segura M, Zheng H, Streptococcus suis sequence type 7 outbreak, Sichuan, China. Emerg Infect Dis. 2006;12:1203–8. DOIPubMedGoogle Scholar
- Yu H, Jing H, Chen Z, Zheng H, Zhu X, Wang H, Human Streptococcus suis outbreak, Sichuan, China. Emerg Infect Dis. 2006;12:914–20. DOIPubMedGoogle Scholar
- Chen C, Tang J, Dong W, Wang C, Feng Y, Wang J, A glimpse of streptococcal toxic shock syndrome from comparative genomics of S. suis 2 Chinese isolates. PLoS ONE. 2007;2:e315. DOIPubMedGoogle Scholar
- Li M, Shen X, Yan J, Han H, Zheng B, Liu D, GI-type T4SS-mediated horizontal transfer of the 89K pathogenicity island in epidemic Streptococcus suis serotype 2. Mol Microbiol. 2011;79:1670–83. DOIPubMedGoogle Scholar
- Feng Y, Shi X, Zhang H, Zhang S, Ma Y, Zheng B, Recurrence of human Streptococcus suis infections in 2007: three cases of meningitis and implications that heterogeneous S. suis 2 circulates in China. Zoonoses Public Health. 2009;56:506–14. DOIPubMedGoogle Scholar
- Feng Y, Zheng F, Pan X, Sun W, Wang C, Dong Y, Existence and characterization of allelic variants of Sao, a newly identified surface protein from Streptococcus suis. FEMS Microbiol Lett. 2007;275:80–8. DOIPubMedGoogle Scholar
- Li M, Wang C, Feng Y, Pan X, Cheng G, Wang J, SalK/SalR, a two-component signal transduction system, is essential for full virulence of highly invasive Streptococcus suis serotype 2. PLoS ONE. 2008;3:e2080 . DOIPubMedGoogle Scholar
Figure
Cite This Article1These authors contributed equally to this article.
Related Links
Table of Contents – Volume 22, Number 6—June 2016
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Youjun Feng, Department of Medical Microbiology and Parasitology, Zhejiang University School of Medicine, Hangzhou, Zhejiang 310058, China; ; or Qinghua Hu, Shenzhen Centre for Disease Control and Prevention, Shenzhen City, Guangdong 518055, China
Top