Volume 22, Number 7—July 2016
Dispatch
Infection with Possible Novel Parapoxvirus in Horse, Finland, 2013
Abstract
A horse in Finland exhibited generalized granulomatous inflammation and severe proliferative dermatitis. After euthanization, we detected poxvirus DNA from a skin lesion sample. The virus sequence grouped with parapoxviruses, closely resembling a novel poxvirus detected in humans in the United States after horse contact. Our findings indicate horses may be a reservoir for zoonotic parapoxvirus.
Parapoxviruses (PPVs) are zoonotic viruses that have been known for centuries to cause contagious pustular skin infections in sheep, goats, and cattle worldwide. These viruses also infect other animals, such as red deer, seals, camels, reindeer, and domestic cats (1,2). In the genus Parapoxvirus, 4 species are currently recognized: Orf virus (ORFV), bovine papular stomatitis virus (BPSV), pseudocowpox virus (PCPV), and parapoxvirus of red deer in New Zealand (PVNZ) (3). In Finland, ORFV has repeatedly been detected in sheep, PCPV in cattle, and ORFV and PCPV in reindeer and humans (4,5). PPVs replicate in epidermal keratinocytes and generally produce pustular lesions at the infection site, which is typically around the mouth, tongue, lips, or teats of mammals. Primary lesions can be severe and proliferative but in uncomplicated cases scab within 1 week and resolve in 4–6 weeks. If the disease is complicated by secondary bacteria, the lesions can become ulcerative and necrotic, delaying healing (6).
All recognized PPV species except PVNZ have been identified in humans. Manifestations of human PPV infections (“farmyard pox”) are typically seen on the hands of persons who had contact with infected ruminants. Recently, Osadebe et al. (7) reported novel poxvirus infections in 2 humans who had contact with domestic animals including horses and donkeys.
In Finland, PPV infections are common in ruminants, but unknown in horses; 3.1% of horses are seropositive for orthopoxviruses (OPV), but such infections appear to be subclinical (8). We describe a severe disease including dermatitis in a horse and identification of possible novel zoonotic parapoxvirus from a skin lesion.
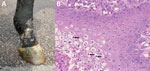
A rapidly progressive disease developed in a 2-year-old Standardbred stallion in Finland; clinical signs were fever, scrotal swelling, and ventral edema (Technical Appendix Figure); multifocal, hard, nodular skin lesions (Figure 1, panel A) and moderately enlarged lymph nodes were also apparent. The horse was apathetic and lost weight despite a good appetite. The attending clinicians suspected generalized lymphoma. However, a biopsy sample taken from nodular skin lesions showed proliferative dermatitis (Table). The horse had secondary immune-mediated hemolytic anemia 1.5 months after onset of disease; because the prognosis was poor, the horse was euthanized in September 2013. The body was received at the University of Helsinki Faculty of Veterinary Medicine (Helsinki, Finland) for a postmortem examination that month.
In necropsy, the horse was found to be thin and poorly muscled. Multifocal, nodular, dry, hard, proliferative lesions in the skin were mainly on the muzzle, lower forelimbs, and ventral abdomen. Moderate edema was present in the abdomen, scrotum, and all limbs. Thickened and hyperemic mucosa in the small intestine, moderately swollen mesenteric lymph nodes, and ascites were visible.
Histologically, the skin lesions were characterized by severe multifocal lymphohistiocytic dermatitis with intraepidermal vesicles caused by marked ballooning degeneration of the stratum granulosum (Figure 1, panel B). Eosinophilic intracytoplasmic inclusion bodies were seen in keratinocytes. Intestinal tissue, lungs, and mesenteric lymph nodes showed chronic, lymphohistiocytic inflammatory changes (Table). Special stains for mycobacteria were negative.
Because the histological findings of the skin samples suggested poxvirus infection, we collected a frozen plain skin sample and slices from formalin-fixed, paraffin-embedded skin, lung, lymph node, and spleen for virological studies. We attempted virus isolation from the skin sample in green monkey and baby hamster kidney cells and saw negative results. DNA was extracted by using the DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany), but no OPV DNA was detectable by real-time PCR (9) (Technical Appendix Table). However, PPV DNA or that of a closely related virus was present in the skin samples: both the Pan-PPV PCR targeting the PPV envelope phospholipase gene (ENV) (11) and the high-GC (guanine-cytosine) pan-pox PCR targeting the large subunit of the poxvirus RNA polymerase gene (RPO147) (10) produced amplicons (Table), although several other primer pairs targeting PPV genes were negative (Technical Appendix Table).
Sequencing of the PCR products showed that the ENV (GenBank accession no. KR863114) and RPO147 (GenBank accession no. KR827441) sequences shared 80%–89% nt and aa identity with other PPVs, depending on the virus species. The RPO147 sequence was 99%–100% identical at nt level and 100% identical at aa level to the sequences of the 2 recent poxvirus isolates (2012_37 and 2013_013 RPO147) from humans in the United States (7). In phylogenetic analyses, the sequences from the horse in this study and from these human patients grouped together, forming a different lineage within the PPVs and separate from other related poxviruses, molluscum contagiosum virus and squirrelpox virus (Figure 2). The equine poxvirus was designated F14.1158H.
Although the skin lesions showed poxvirus infection, formalin-fixed samples from internal organs contained no viral inclusion bodies and were negative for PPV by PCR. This finding is in accordance with the fact that PPVs are specialized to replicate in the highly specific immune environment of skin (13). Further investigations are required to show whether the poxvirus caused the generalized infection in addition to dermatitis.
The owner, breeder, and trainers of the horses on the farm where this horse became ill were unaware of any other animal or zoonotic cases in the premises and disclosed no contact between the horse and ruminants. The horse had lived in contact with many horses and several dogs and cats in 3 locations in southern parts of western and eastern Finland before being transferred to the last training stable. A few months before onset of clinical signs, the horse had been trained at a farm where cows had been kept 25 years earlier. During the illness, the horse lived in a stable with 17 horses, shared corrals and equipment, and had muzzle contact with 2 horses in adjacent stalls. Despite the direct and indirect contacts, all other horses, the 3 caretakers, and the trainer remained asymptomatic.
We report a clinical equine infection with a novel poxvirus in Finland. The infection is at least of dermatitic relevance for horses, and veterinary awareness is needed. The sequence analysis based on conserved genes revealed a close relationship between this isolate and recent poxvirus isolates from humans with horse contact in the United States (7). Although sequence data are limited and the geographic distance between this equine case and the recent cases in humans is remote, the close genetic relatedness suggests that horses have a possible role as reservoir or vector of an emerging zoonotic poxvirus, necessitating medical awareness and emphasizing the importance of the One Health approach (https://www.onehealthcommission.org/). The horse as an origin for zoonoses is not uncommon: as many as 58% of emerging zoonotic pathogens infect ungulates (14). As for cowpox virus, horse and human may be infected from a common source, such as rodents, and not necessarily from each other. This case appeared sporadic and not very contagious, and the transmission route remained unresolved. Further studies are needed to elucidate ecology, epidemiology, prevalence, and possible zoonotic transmission.
As our limited sequence analysis suggests, the virus we detected is most closely related to PPVs and may merit being classified as a new Parapoxvirus species. However, many of the established PPV primer pairs did not produce PCR product, which suggests that the virus is different from the established PPV species and may represent a new poxvirus genus. More sequence data are needed to validate the taxonomic classification of the equine poxvirus. In conclusion, our results provide further evidence that horses are a possible source of the new poxvirus infection recently observed in humans.
Dr. Airas is a senior lecturer in the field of veterinary pathology and parasitology, Faculty of Veterinary Medicine, University of Helsinki, Helsinki, Finland. Her main research interests are pathology of domestic animals and parasitology, especially Trichinella spp.
Acknowledgment
We thank Sanna Malkamäki for her help with the necropsy, as well as Laura Mannonen, Irja Luoto, and Kirsi Aaltonen for their generous help with virological studies. We also thank Maija Huttunen for excellent technical assistance, Michael Hewetson for providing the pictures of the horse, and the owner, breeder, and trainers of the horse patient for their cooperation.
References
- Essbauer S, Pfeffer M, Meyer H. Zoonotic poxviruses. Vet Microbiol. 2010;140:229–36 .DOIPubMedGoogle Scholar
- Fairley RA, Whelan EM, Pesavento PA, Mercer AA. Recurrent localized cutaneous parapoxviruses infection in three cats. N Z Vet J. 2008;56:196–201 .DOIPubMedGoogle Scholar
- Skinner MA, Buller RM, Damon IK, Lefkowitz EJ, McFadden G, Mc Innes CJ, 2012. Poxviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus taxonomy: classification and nomenclature of viruses. Ninth report of the international committee on taxonomy of viruses. San Diego: Elsevier Academic Press, 2012. p. 291–309.
- Tikkanen MK, McInnes CJ, Mercer AA, Buttner M, Tuimala J, Hirvela-Koski V, Recent isolates of parapoxvirus of Finnish reindeer (Rangifer tarandus tarandus) are closely related to bovine pseudocowpox virus. J Gen Virol. 2004;85:1413–8. DOIPubMedGoogle Scholar
- Hautaniemi M, Vaccari F, Scagliarini A, Laaksonen S, Huovilainen A, McInnes CJ. Analysis of deletion within the reindeer pseudocowpoxvirus genome. Virus Res. 2011;160:326–32. DOIPubMedGoogle Scholar
- Osadebe LU, Manhiram K, McCollum AM, Li Y, Emerson GL, Gallardo-Romero NF, Novel poxvirus infection in 2 patients from the United States. Clin Infect Dis. 2015;60:195–202. DOIPubMedGoogle Scholar
- Kinnunen PM. Detection and epidemiology of cowpox and Borna disease virus infections [dissertation]. Helsinki (Finland): University of Helsinki; 2011 [cited 2015 Oct 3]. https://helda.helsinki.fi/handle/10138/27343
- Putkuri N, Piiparinen H, Vaheri A, Vapalahti O. Detection of human orthopoxvirus infections and differentiation of smallpox virus with real-time PCR. J Med Virol. 2009;81:146–52. DOIPubMedGoogle Scholar
- Li Y, Meyer H, Zhao H, Damon IK. GC content-based pan-pox universal PCR assays for poxvirus detection. J Clin Microbiol. 2010;48:268–76. DOIPubMedGoogle Scholar
- Inoshima Y, Morooka A, Sentsui H. Detection and diagnosis of parapoxvirus by the polymerase chain reaction. J Virol Methods. 2000;84:201–8. DOIPubMedGoogle Scholar
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30:2725–9. DOIPubMedGoogle Scholar
- Fleming SB, Wise LM, Mercer AA. Molecular genetic analysis of orf virus: a poxvirus that has adapted to skin. Viruses. 2015;7:1505–39. DOIPubMedGoogle Scholar
- Cleaveland S, Laurenson MK, Taylor LH. Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Philos Trans R Soc Lond B Biol Sci. 2001;356:991–9 . DOIPubMedGoogle Scholar
Figures
Table
Cite This Article1Current affiliation: Turku University Hospital, Turku, Finland.
2Current affiliation: Finnish Food Safety Authority Evira, Helsinki, Finland.
Table of Contents – Volume 22, Number 7—July 2016
| EID Search Options |
|---|
|
|
|
|
|
|
![Thumbnail of Phylogenetic analyses of sequences amplified from skin lesion of horse infected with possible novel parapoxvirus, Finland, 2013 (poxvirus variant F14.1158H), and other poxviruses. Trees were generated by using the neighbor-joining method in MEGA 6 software (http://www.megasoftware.net) (12), based on A) 184 aa of envelope phospholipase gene and B) 195 aa of viral RNA polymerase gene RP0147. GenBank accession numbers for sequences used in the analyses: JF773701 (Orf virus [ORFV] F07.](/eid/images/15-1636-F2-tn.jpg)
Please use the form below to submit correspondence to the authors or contact them at the following address:
Niina Airas, Faculty of Veterinary Medicine, Department of Veterinary Biosciences, Veterinary Pathology, PO Box 66, Agnes Sjöbergin katu 2, 00014 University of Helsinki, Helsinki, Finland
Top