Volume 25, Number 5—May 2019
Dispatch
Bombali Virus in Mops condylurus Bat, Kenya
Abstract
Bombali virus (genus Ebolavirus) was identified in organs and excreta of an Angolan free-tailed bat (Mops condylurus) in Kenya. Complete genome analysis revealed 98% nucleotide sequence similarity to the prototype virus from Sierra Leone. No Ebola virus–specific RNA or antibodies were detected from febrile humans in the area who reported contact with bats.
The virus family Filoviridae is divided into 5 genera: Cuevavirus, Marburgvirus, Ebolavirus, Striavirus, and Thamnovirus (https://talk.ictvonline.org/taxonomy). Six distinct Ebola viruses have been described; 4 are known to cause human disease (1,2). These include highly lethal pathogens capable of producing large outbreaks, namely Bundibugyo, Sudan, and Zaire Ebola viruses, the last responsible for the devastating 2013–2016 outbreak in West Africa and an ongoing extended outbreak in the Democratic Republic of the Congo (1,3,4). Although the natural reservoirs of Ebola viruses remain unconfirmed, considerable evidence supports a role for bat species, particularly fruit bats, analogous to findings implicating Rousettus aegypticus fruit bats as a reservoir for Marburg virus (1,5,6).
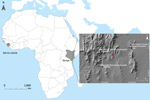
The most recent Ebola virus to be identified is named Bombali virus (BOMV) and was reported in August 2018 in mouth and fecal swabs collected from free-tailed insectivorous bat species (family Molossidae) Mops condylurus and Chaerephon pumilus in Sierra Leone (2). Although BOMV is not known to infect humans, its envelope glycoprotein shares the same NPC1 receptor as other filoviruses and is capable of mediating BOMV pseudotype virus entry into human cells (2). We describe the presence of BOMV in tissues and excreta of an Angolan free-tailed bat (M. condylurus) captured near the Taita Hills in southeastern Kenya, the easternmost distributional range of this bat species (7), >5,500 km from the original BOMV identification site in Sierra Leone (Figure 1). We also screened human serum samples collected from febrile patients in the Taita Hills area for markers of BOMV infection.
We identified BOMV in an adult female bat (B241) by reverse transcription PCR and next-generation sequencing. This bat was captured along with 15 others in mist nets in savannah habitat near a small river in May 2018; only this bat was BOMV positive; (6% prevalence). Viral RNA was present in lung, spleen, liver, heart, intestine, mouth swab, and fecal samples but absent from the brain, kidney, urine, and a few fleas found on the bat; viral loads were especially high in the lung (Appendix). These tissue-positive findings confirm that BOMV can infect M. condylurus and is not an artifact of its insect diet, which could not be discounted from the previous analysis on the basis of mouth and fecal swabs (2). We also screened lung samples of sympatric C. pumilus bats (n = 13) and other bat species (Appendix Table 2) captured from the same area in February 2016 and May 2018; all were negative for BOMV RNA. Serologic analysis revealed antibodies against Ebola virus in the blood of the tissue-positive bat (Appendix Figure), but specific antibodies were not found in blood from the other bats (Appendix).
Our tissue-positive findings provide a strong host association between BOMV and M. condylurus bats; it is possible that BOMV–positive findings from other bat species result from local spillover or contamination. Moreover, phylogenetic analysis of the full BOMV genome from the bat lung revealed 98% nucleotide sequence similarity with the prototype reported in Sierra Leone (GenBank accession no. MK340750) (Figure 2). Considering the high sequence similarity between the 2 locations and that M. condylurus bats, like most insectivorous bats, are believed to travel only short distances (8), BOMV is likely to be distributed throughout much of sub-Saharan Africa (7). However, further monitoring of M. condylurus and C. pumilus bats and other sympatric species across Africa is required to support this hypothesis.
Because M. condylurus bats commonly roost in human structures, such as house roofs (8,9), human exposure to this species is more likely than for many other bat species. Therefore, we screened for markers of human infection with BOMV by studying serum samples collected from febrile patients who sought treatment at clinics in the Taita Hills area during April–August 2016. Clinics are located in the surrounding areas, all within 15 km of the BOMV–infected bat collection site (Figure 1). We screened patients for filovirus RNA (n = 81) and Ebola virus–specific IgG (n = 250) by an immunofluorescence assay using Zaire Ebola virus VP40–transfected VeroE6 cells as antigen (Appendix). Many samples, including all those screened for filovirus RNA, were from patients who reported contact with bats in the home or workplace. We found no evidence of filovirus infection by either screening method, providing no support that BOMV easily infects humans or is a common cause of febrile illness in the area. Ongoing surveillance is nonetheless necessary, and we cannot exclude the possibility that BOMV was a recent introduction to the Taita Hills area.
Our results markedly expand the distributional range of this new Ebola virus to eastern Africa and confirm the M. condylurus bat as a competent host. Like Goldstein et al. (2), we stress that the virus is not known to infect humans, a premise supported by our screening of febrile patients in the Taita Hills area. Potential efforts to eradicate bats are unwarranted and may jeopardize their crucial ecosystem roles and human health (10,11).
Dr. Forbes is a disease ecologist and assistant professor at the Department of Biological Sciences, University of Arkansas. His research interests include the maintenance and transmission of rodentborne and batborne zoonotic pathogens in nature and the effects of anthropogenic environmental changes on these processes.
Acknowledgments
We thank Joni Uusitalo and Michael Bartonjo for fieldwork assistance; Johanna Martikainen, Mira Utriainen, and Fathiah Zakham for laboratory assistance; and Ruut Uusitalo for preparing the figure map. We also thank the clinicians who recruited patients for the human study and the staff at the University of Helsinki Taita Research Station and its director, Petri Pellikka, for facilitating this work.
This research was supported by the Finnish Cultural Foundation, the Jenny and Antti Wihuri Foundation, the Academy of Finland (grant no. 318726), Helsinki University Hospital Funds, and the Jane and Aatos Erkko Foundation. A.J.J., S.A., L.L., A.M., O.V., and T.S. are part of the EbolaMoDRAD consortium, which has received funding to develop diagnostic methods for filoviruses from the Innovative Medicine Initiative 2 Joint Undertaking under grant agreement no. 115843. This Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation program and the European Federation of Pharmaceutical Industries and Associations (EFPIA). Bat trapping and sample collections were carried out under permits from the National Commission for Science, Technology, and Innovation (permit no. NACOSTI/P/18/76501/22243) and the Kenya Wildlife Service (permit no. KWS/BRM/500). The Finnish Food Safety Authority (Evira) approved the importation of samples (permit nos. 4250/0460/2016 and 2809/0460/2018). Kenyatta National Hospital and the University of Nairobi Ethics and Research Committee approved the human study (permit no. P707/11/2015).
References
- Olival KJ, Hayman DTS. Filoviruses in bats: current knowledge and future directions. Viruses. 2014;6:1759–88. DOIPubMedGoogle Scholar
- Goldstein T, Anthony SJ, Gbakima A, Bird BH, Bangura J, Tremeau-Bravard A, et al. The discovery of Bombali virus adds further support for bats as hosts of ebolaviruses. Nat Microbiol. 2018;3:1084–9. DOIPubMedGoogle Scholar
- Piot P, Muyembe JJ, Edmunds WJ. Ebola in west Africa: from disease outbreak to humanitarian crisis. Lancet Infect Dis. 2014;14:1034–5. DOIPubMedGoogle Scholar
- Nkengasong JN, Onyebujoh P. Response to the Ebola virus disease outbreak in the Democratic Republic of the Congo. Lancet. 2018;391:2395–8. DOIPubMedGoogle Scholar
- Towner JS, Amman BR, Sealy TK, Carroll SA, Comer JA, Kemp A, et al. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 2009;5:e1000536. DOIPubMedGoogle Scholar
- Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, et al. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–6. DOIPubMedGoogle Scholar
- Happold M. Tadarida condylura Angolan free-tailed bat. In: Happold M, Happold D, editors. Mammals of Africa (hedgehogs, shrews, and bats). Vol. 4. London: Bloomsbury; 2013. p. 505–7.
- Noer CL, Dabelsteen T, Bohmann K, Monadjem A. Molossid bats in an African agro-ecosystem select sugarcane fields as foraging habitat. Afr Zool. 2012;47:1–11. DOIGoogle Scholar
- Bronrier GN, Maloney SK, Buffenstein R. Survival tactics within thermally-challenging roosts: heat tolerance and cold sensitivity in the Angolan free-tailed bat, Mops condylurus. S Afr Zool. 1999;34:1–10. DOIGoogle Scholar
- Kunz TH, Braun de Torrez E, Bauer D, Lobova T, Fleming TH. Ecosystem services provided by bats. Ann N Y Acad Sci. 2011;1223:1–38. DOIPubMedGoogle Scholar
- Amman BR, Nyakarahuka L, McElroy AK, Dodd KA, Sealy TK, Schuh AJ, et al. Marburgvirus resurgence in Kitaka Mine bat population after extermination attempts, Uganda. Emerg Infect Dis. 2014;20:1761–4. DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: April 01, 2019
1Current affiliation: University of Arkansas, Fayetteville, Arkansas, USA.
Table of Contents – Volume 25, Number 5—May 2019
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Kristian M. Forbes, University of Arkansas, Department of Biological Sciences, SCEN Bldg, 850 W Dickson St, Fayetteville, AR 16801, USA
Top