Volume 30, Number 5—May 2024
Dispatch
Antigenic Characterization of Novel Human Norovirus GII.4 Variants San Francisco 2017 and Hong Kong 2019
Abstract
Norovirus is a major cause of acute gastroenteritis; GII.4 is the predominant strain in humans. Recently, 2 new GII.4 variants, Hong Kong 2019 and San Francisco 2017, were reported. Characterization using GII.4 monoclonal antibodies and serum demonstrated different antigenic profiles for the new variants compared with historical variants.
Norovirus is a major cause of acute gastroenteritis (1). Over past decades, variants of the predominant genotype, genotype 4 (GII.4), have continuously emerged to escape immunity (2–5). Since 2012, the Sydney 2012 variant has predominated worldwide (6). In 2019, GII.4 noroviruses that did not cluster with any known variants were reported circulating in different countries as early as 2016 (7,8). Based on phylogenetic clustering and number of mutations on major capsid viral proteins (VP1), the variant was classified GII.4 Hong Kong 2019 (7). The new variant caused no large outbreaks and did not eclipse the predominance of Sydney 2012. Another recently reported unique group of GII.4 noroviruses, the San Francisco 2017 variant, was retrospectively detected circulating during 2017–2022 (9). Both variants showed multiple mutations on major antigenic sites, including a single amino acid insertion next to the antigenic site A in San Francisco 2017 (7,9,10). We characterize the antigenicity of these 2 new variants using panels of GII.4 mouse monoclonal antibodies (mAbs) and hyperimmune serum developed against historical GII.4 variants (11,12).
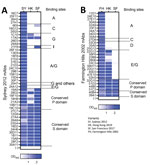
To determine the cross-reactivity of the 2 new variants with previously circulating variants, we produced virus-like particles (VLPs) for Hong Kong 2019 (GenBank accession no.: MN400355) and San Francisco 2017 (GenBank accession no. MW506849) viruses. We performed ELISA by using mAbs developed against Sydney 2012 virus and the newly developed VLPs (11). Results demonstrated that the Hong Kong 2019 VLPs bound to most of the mAbs mapping to conserved sites from protruding (P) and shell (S) domains of the VP1, but only bound to 2/25 mAbs that mapped to variable antigenic sites and showed histo-blood group antigen (HBGA) blockade activity (Figure 1, panel A) (11). Those results were expected because mutational analyses showed that the Hong Kong 2019 viruses present multiple mutations on variable antigenic sites (Appendix) (7). The loss of binding of 3 cross-reactive mAbs that mapped to the P domain could be explained by unique mutations on the conserved sites (Appendix). VLPs from the San Francisco 2017 variant bound to all mAbs mapping to conserved sites of the VP1, but only to 4 mAbs that mapped to variable antigenic sites. Based on previous observations, alanine on positions 356, 359, or both, play a role in binding to mAbs 1C10 and 17A5 (11). Thus, alanine on those positions could explain the binding of mAbs to Hong Kong 2019 and San Francisco 2017 VLPs. Other mAbs mapping to the antigenic site G, 26E5 and 29A9, seem to require residues from antigenic site A (11). Mutations on antigenic site A in San Francisco 2017 could therefore result in loss of binding of these mAbs regardless of similarity to antigenic site G on the Sydney 2012 variant. The 6E6 mAb, mapping to antigenic site C, was previously reported to cross-react weakly to Farmington Hills 2002 variant (11), which has 3 mutations compared with Sydney 2012. The San Francisco 2017 presented 4 mutations compared with Sydney 2012; the Hong Kong 2019 variant had 6 mutations on that site, explaining the differential binding of this mAb. Similarly, sequence differences on antigenic site I could explain the lack of binding of mAb 21F8 to Hong Kong 2019 VLPs.
Because Hong Kong 2019 and San Francisco 2017 present evolutionary convergence and share similar residues on several of the antigenic sites compared with the Farmington Hills 2002 variant (Appendix), we also tested those strains with mAbs developed against this ancestral variant (Figure 1, panel B). Both VLPs showed reactivity with all mAbs binding to conserved epitopes. As expected based on sequence similarity, Hong Kong 2019 showed reactivity only with mAbs mapping on antigenic site E/G. San Francisco 2017 was negative to all mAbs mapping to variable sites, including antigenic site A, which presented only 3 mutations from Farmington Hills 2002 VLPs. Those data indicate that either a small number of changes are sufficient to abrogate binding of all 9 A-mapping mAbs or that the insertion near the antigenic site A has a major influence on the characteristics of this antigenic site.
To further characterize the antigenicity of these new variants, we tested the HBGA blocking activity of serum from mice immunized with VLPs from historical variants (Figure 2, panels A, B) (12), including the currently circulating Sydney 2012, the ancestral Farmington Hills 2002, and genetically or phylogenetically related variants: Osaka 2007 for Hong Kong 2019 viruses (7), and New Orleans 2009 and Apeldoorn 2007 for San Francisco 2017 viruses (9) (Appendix). The Hong Kong 2019 VLPs presented weak cross-blockade reactivity with the serum raised against all 3 viruses (mean 50% effective concentration = 118.5 for Sydney 2012, 116.9 for Farmington Hills 2002, and 87.6 for Osaka 2007 serum), with >12-fold differences compared with their homologous VLPs (Figure 2, panel A). That result was consistent with data from children with the fewest previous norovirus infections (10), supporting minimal cross-reactivity of Hong Kong 2019 to previous variants. In contrast, the San Francisco 2017 VLPs did not show cross-blockade reactivity with any of the serum samples tested from pandemic variants (Figure 2, panel B), including Sydney 2012 and Farmington Hills 2002, that shared similar sequences on the antigenic sites E/G (Sydney 2012) and A (Farmington Hills 2002) (Appendix). The only exception for cross-reactivity was with the serum raised against Apeldoorn 2007, which showed moderate cross-blockade activity with the San Francisco 2017 (50% effective concentration = 309.2, a 5-fold difference compared with homologous VLPs). Of note, GII.4 Apeldoorn 2007 and San Francisco 2017 share the same motif on the antigenic site D (Appendix). Thus, that cross-reactivity might be explained by antibodies mapping on the antigenic site D from Apeldoorn 2007 variant.
Our data indicate that both Hong Kong 2019 and San Francisco 2017 variants present distinct antigenic profiles, yet both viruses have been circulating for >7 years without causing large outbreaks globally. Multiple examples of antigenically distinct noroviruses that spread worldwide without causing large outbreaks exist. Minor variants such as Osaka 2007 and Apeldoorn 2007 showed distinct antigenic profiles to variants that circulated before their emergence (12). Those variants caused local outbreaks and spread to multiple countries, but none predominated at the global level (13). Therefore, changes in antigenicity might not be the only factor determining the epidemic potential of noroviruses. Indeed, specific HBGA binding profiles were associated with emerging noroviruses (14,15). The new GII.4 variants bound to porcine gastric mucin III and human saliva, as did other current and archival variants (9,10) (Figure 2, panel C), suggesting that impairment of binding to HBGA did not cause the lower circulation of these viruses.
In conclusion, these 2 new norovirus variants are antigenically distinct from previously circulating variants. Whether these variants will predominate or are examples of the subdued circulation of minor norovirus variants remains to be determined. To prepare for future pandemics, we must delineate the factors that determine the overall fitness and predominance of GII.4 noroviruses, including but not limited to replication kinetics, pathogenicity, HBGA binding spectrum, and epidemiologic confounder.
Dr. Tohma is a visiting associate at the Division of Viral Products, Food and Drug Administration, Silver Spring, Maryland, USA. His research interests include viral genetic and antigenic evolution, epidemiology, and norovirus vaccine design.
Acknowledgments
We thank Yamei Gao for her technical support on virus-like particle production.
Financial support for this work was provided by the US Food and Drug Administration intramural funds (program no. Z01 BK 04012 LHV–G.I.P). K.A.P. and M.L. are recipients of a CBER/FDA-sponsored Oak Ridge Institute for Science and Education (ORISE) fellowship.
References
- Lopman BA, Steele D, Kirkwood CD, Parashar UD. The vast and varied global burden of norovirus: prospects for prevention and control. PLoS Med. 2016;13:
e1001999 . DOIPubMedGoogle Scholar - Lindesmith LC, Costantini V, Swanstrom J, Debbink K, Donaldson EF, Vinjé J, et al. Emergence of a norovirus GII.4 strain correlates with changes in evolving blockade epitopes. J Virol. 2013;87:2803–13. DOIPubMedGoogle Scholar
- Lindesmith LC, Donaldson EF, Lobue AD, Cannon JL, Zheng DP, Vinje J, et al. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 2008;5:
e31 . DOIPubMedGoogle Scholar - Siebenga JJ, Lemey P, Kosakovsky Pond SL, Rambaut A, Vennema H, Koopmans M. Phylodynamic reconstruction reveals norovirus GII.4 epidemic expansions and their molecular determinants. PLoS Pathog. 2010;6:
e1000884 . DOIPubMedGoogle Scholar - Tohma K, Lepore CJ, Gao Y, Ford-Siltz LA, Parra GI. Population genomics of GII.4 noroviruses reveal complex diversification and new antigenic sites involved in the emergence of pandemic strains. MBio. 2019;10:10. DOIPubMedGoogle Scholar
- Parra GI, Tohma K, Ford-Siltz LA, Eguino P, Kendra JA, Pilewski KA, et al. Minimal antigenic evolution after a decade of norovirus GII.4 Sydney_2012 circulation in humans. J Virol. 2023;97:
e0171622 . DOIPubMedGoogle Scholar - Chan MC, Roy S, Bonifacio J, Zhang LY, Chhabra P, Chan JCM, et al.; for NOROPATROL2. for NOROPATROL2. Detection of norovirus variant GII.4 Hong Kong in Asia and Europe, 2017–2019. Emerg Infect Dis. 2021;27:289–93. DOIPubMedGoogle Scholar
- Chuchaona W, Chansaenroj J, Puenpa J, Khongwichit S, Korkong S, Vongpunsawad S, et al. Human norovirus GII.4 Hong Kong variant shares common ancestry with GII.4 Osaka and emerged in Thailand in 2016. PLoS One. 2021;16:
e0256572 . DOIPubMedGoogle Scholar - Chhabra P, Tully DC, Mans J, Niendorf S, Barclay L, Cannon JL, et al. Emergence of novel norovirus GII.4 variant. Emerg Infect Dis. 2024;30:163–7. DOIPubMedGoogle Scholar
- Lindesmith LC, Boshier FAT, Brewer-Jensen PD, Roy S, Costantini V, Mallory ML, et al. Immune imprinting drives human norovirus potential for global spread. MBio. 2022;13:
e0186122 . DOIPubMedGoogle Scholar - Tohma K, Ford-Siltz LA, Kendra JA, Parra GI. Dynamic immunodominance hierarchy of neutralizing antibody responses to evolving GII.4 noroviruses. Cell Rep. 2022;39:
110689 . DOIPubMedGoogle Scholar - Kendra JA, Tohma K, Ford-Siltz LA, Lepore CJ, Parra GI. Antigenic cartography reveals complexities of genetic determinants that lead to antigenic differences among pandemic GII.4 noroviruses. Proc Natl Acad Sci U S A. 2021;118:
e2015874118 . DOIPubMedGoogle Scholar - Kendra JA, Tohma K, Parra GI. Global and regional circulation trends of norovirus genotypes and recombinants, 1995-2019: A comprehensive review of sequences from public databases. Rev Med Virol. 2022;32:
e2354 . DOIPubMedGoogle Scholar - Shanker S, Choi JM, Sankaran B, Atmar RL, Estes MK, Prasad BV. Structural analysis of histo-blood group antigen binding specificity in a norovirus GII.4 epidemic variant: implications for epochal evolution. J Virol. 2011;85:8635–45. DOIPubMedGoogle Scholar
- Zhang XF, Huang Q, Long Y, Jiang X, Zhang T, Tan M, et al. An outbreak caused by GII.17 norovirus with a wide spectrum of HBGA-associated susceptibility. Sci Rep. 2015;5:17687. DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: April 17, 2024
Table of Contents – Volume 30, Number 5—May 2024
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Kentaro Tohma, Center for Biologics Evaluation and Research, US Food and Drug Administration, 10903 New Hampshire Ave, Bldg 52/72, Rm 1376, Silver Spring, MD 20993, USA
Top