Volume 30, Number 5—May 2024
Dispatch
Antigenic Characterization of Novel Human Norovirus GII.4 Variants San Francisco 2017 and Hong Kong 2019
Figure 1
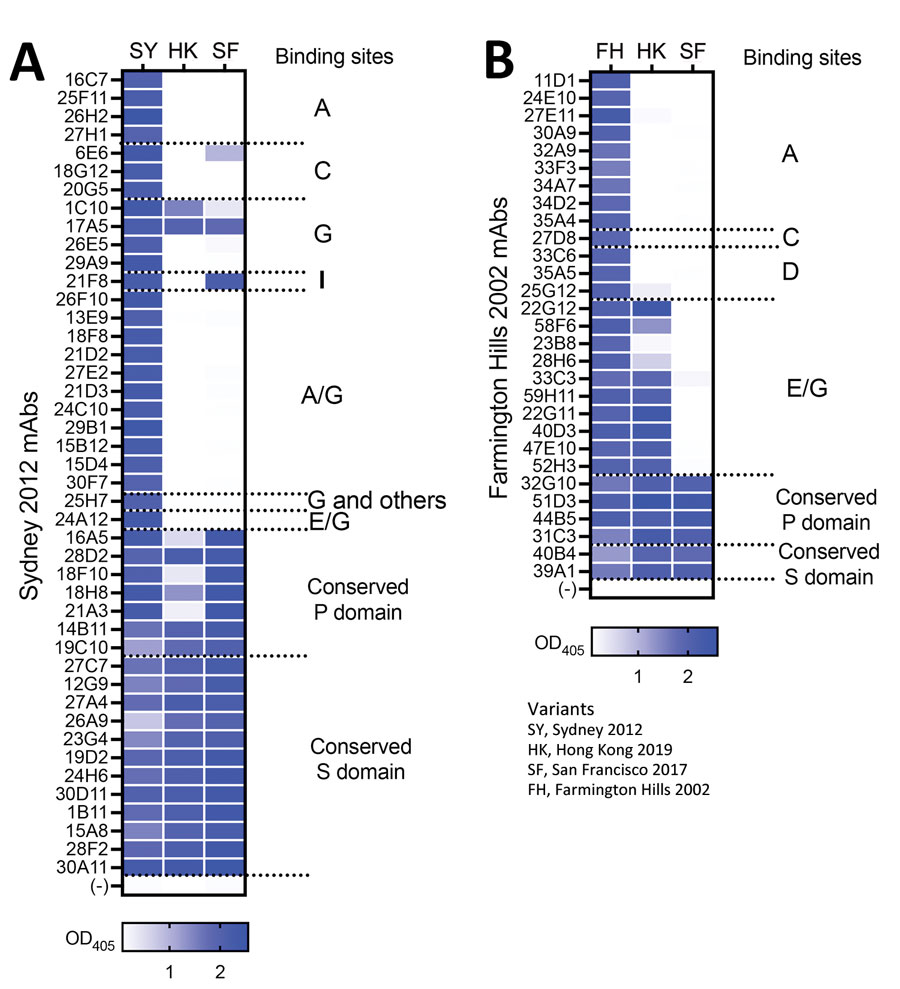
Figure 1. Monoclonal antibodies raised against 2 major GII.4 variants in a study of novel human norovirus GII.4 variants, San Francisco 2017 and Hong Kong 2019. A) Sydney 2012 mAb panel; B) Farmington Hills 2002 mAb panel. The heatmaps indicate ELISA binding strength (OD405 values) of individual mAbs against virus-like particles from GII.4 Hong Kong 2019 and San Francisco 2017. Antibodies indicate minimal cross-reactivity between new and previously described variants. The binding sites of the mAbs were characterized in a previous study (11). mAbs, monoclonal antibodies; OD405, optical density at 405 nm; P, protruding; S, shell.
References
- Lopman BA, Steele D, Kirkwood CD, Parashar UD. The vast and varied global burden of norovirus: prospects for prevention and control. PLoS Med. 2016;13:
e1001999 . DOIPubMedGoogle Scholar - Lindesmith LC, Costantini V, Swanstrom J, Debbink K, Donaldson EF, Vinjé J, et al. Emergence of a norovirus GII.4 strain correlates with changes in evolving blockade epitopes. J Virol. 2013;87:2803–13. DOIPubMedGoogle Scholar
- Lindesmith LC, Donaldson EF, Lobue AD, Cannon JL, Zheng DP, Vinje J, et al. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 2008;5:
e31 . DOIPubMedGoogle Scholar - Siebenga JJ, Lemey P, Kosakovsky Pond SL, Rambaut A, Vennema H, Koopmans M. Phylodynamic reconstruction reveals norovirus GII.4 epidemic expansions and their molecular determinants. PLoS Pathog. 2010;6:
e1000884 . DOIPubMedGoogle Scholar - Tohma K, Lepore CJ, Gao Y, Ford-Siltz LA, Parra GI. Population genomics of GII.4 noroviruses reveal complex diversification and new antigenic sites involved in the emergence of pandemic strains. MBio. 2019;10:10. DOIPubMedGoogle Scholar
- Parra GI, Tohma K, Ford-Siltz LA, Eguino P, Kendra JA, Pilewski KA, et al. Minimal antigenic evolution after a decade of norovirus GII.4 Sydney_2012 circulation in humans. J Virol. 2023;97:
e0171622 . DOIPubMedGoogle Scholar - Chan MC, Roy S, Bonifacio J, Zhang LY, Chhabra P, Chan JCM, et al.; for NOROPATROL2. for NOROPATROL2. Detection of norovirus variant GII.4 Hong Kong in Asia and Europe, 2017–2019. Emerg Infect Dis. 2021;27:289–93. DOIPubMedGoogle Scholar
- Chuchaona W, Chansaenroj J, Puenpa J, Khongwichit S, Korkong S, Vongpunsawad S, et al. Human norovirus GII.4 Hong Kong variant shares common ancestry with GII.4 Osaka and emerged in Thailand in 2016. PLoS One. 2021;16:
e0256572 . DOIPubMedGoogle Scholar - Chhabra P, Tully DC, Mans J, Niendorf S, Barclay L, Cannon JL, et al. Emergence of novel norovirus GII.4 variant. Emerg Infect Dis. 2024;30:163–7. DOIPubMedGoogle Scholar
- Lindesmith LC, Boshier FAT, Brewer-Jensen PD, Roy S, Costantini V, Mallory ML, et al. Immune imprinting drives human norovirus potential for global spread. MBio. 2022;13:
e0186122 . DOIPubMedGoogle Scholar - Tohma K, Ford-Siltz LA, Kendra JA, Parra GI. Dynamic immunodominance hierarchy of neutralizing antibody responses to evolving GII.4 noroviruses. Cell Rep. 2022;39:
110689 . DOIPubMedGoogle Scholar - Kendra JA, Tohma K, Ford-Siltz LA, Lepore CJ, Parra GI. Antigenic cartography reveals complexities of genetic determinants that lead to antigenic differences among pandemic GII.4 noroviruses. Proc Natl Acad Sci U S A. 2021;118:
e2015874118 . DOIPubMedGoogle Scholar - Kendra JA, Tohma K, Parra GI. Global and regional circulation trends of norovirus genotypes and recombinants, 1995-2019: A comprehensive review of sequences from public databases. Rev Med Virol. 2022;32:
e2354 . DOIPubMedGoogle Scholar - Shanker S, Choi JM, Sankaran B, Atmar RL, Estes MK, Prasad BV. Structural analysis of histo-blood group antigen binding specificity in a norovirus GII.4 epidemic variant: implications for epochal evolution. J Virol. 2011;85:8635–45. DOIPubMedGoogle Scholar
- Zhang XF, Huang Q, Long Y, Jiang X, Zhang T, Tan M, et al. An outbreak caused by GII.17 norovirus with a wide spectrum of HBGA-associated susceptibility. Sci Rep. 2015;5:17687. DOIPubMedGoogle Scholar
Page created: February 21, 2024
Page updated: April 24, 2024
Page reviewed: April 24, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.