Volume 31, Number 2—February 2025
Dispatch
Bacteremia and Community-Acquired Pneumonia Caused by Pantoea stewartii Subspecies indologenes, Australia
Cite This Article
Citation for Media
Abstract
We report infection with the phytopathogen Pantoea stewartii subspecies indologenes in a macadamia farmer from southeast Queensland, Australia. The patient had bloodstream infection and pneumonia develop after an unidentified inoculation event. Investigation determined that the most likely mode of transmission was inhalation from an environmental source on the farm.
Pantoea species are ubiquitous bacteria in both terrestrial and aquatic environments and have been isolated in animals, insects, and humans, although most of the 31 recognized species (1) are associated with plants (2). Pantoea species previously reported in human infections include P. agglomerans, P. ananatis, P. brenneri, P. calida, P. conspicua, P. dispersa, P. eucrina, and P. septica; P. agglomerans has been the most common (2).
P. stewartii, the cause of Stewart’s wilt in sweet corn and maize, was first discovered in the late 1890s. Researchers proposed 2 subspecies in 1993 based on host range: stewartii and indologenes (3). Unlike subspecies stewartii, subspecies indologenes is nonpathogenic to corn, instead causing disease in other agronomically significant crops, such as foxtail millet, pearl millet, and onions. Because of the risk this organism (particularly subspecies stewartii) poses to economically critical crops, many countries classify P. stewartii as a quarantine organism (4,5).
In 2022, researchers reported infection with P. stewartii in a human, initially identifying the species with moderate confidence as P. septica but then recategorizing the species designation based on 16S ribosomal RNA gene sequencing (1). We report phytopathogen P. stewartii subsp. indologenes infection in a macadamia farmer from southeast Queensland, Australia.
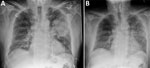
In summer 2021, an 82-year-old man sought care at the emergency department of Sunshine Coast University Hospital (Birtinya, QLD, Australia), for sudden onset of fever, myalgia, arthralgia, nonproductive cough, and shortness of breath. The patient had recently moved to a macadamia farm in the Sunshine Coast hinterland and had previously resided on a pineapple farm in the same region for several decades. He reported no overseas travel history, no preceding history of inoculating injury from plant material, no recent skin wounds or infections, and no direct zoonotic contacts. Chest radiograph (Figure 1, panel A) identified moderately severe community-acquired multilobar pneumonia, with dense consolidation in the left upper lobe. The patient deteriorated rapidly because of septic shock, necessitating intubation and ventilation, and was transferred to the intensive care unit on day 1 of admission.
We observed lactose-fermenting, nonmucoid, yellow-pigmented colonies cultivated on MacConkey and horse blood agars (Figure 2) from blood culture (isolate SCHI0154.S.1), tracheal aspirate, and bronchial washing specimens. We determined the colonies to be catalase positive, oxidase negative, and spot indole positive. VITEK MS matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (bioMérieux, https://www.biomerieux.com) revealed P. ananatis with 99.9% probability. The VITEK GN ID card identified the respiratory isolates as Pantoea sp. with 95% probability. SCHI0154.S.1 was resistant to ampicillin (MIC = 16 mg/L) and cephazolin (MIC ≥64 mg/L) but susceptible to amoxicillin/clavulanate (MIC ≤2 mg/L), ceftriaxone (MIC ≤1 mg/L), ciprofloxacin (MIC ≤0.25 mg/L), gentamicin (MIC ≤1 mg/L), meropenem (MIC ≤0.25 mg/L), piperacillin/tazobactam (MIC = 2 mg/L), and trimethoprim/sulfamethoxazole (MIC ≤20 mg/L).
We treated the patient with intravenous amoxicillin/clavulanate (2.2 g/8 h) for a total of 10 days and successfully extubated him on day 7. Chest radiograph opacification on day 12 showed improvements (Figure 1, panel B). At outpatient follow-up, the patient had reached full recovery.
To permit accurate speciation and genetic comparison with other Pantoea isolates, we compared Illumina NovaSeq 2 × 150bp whole-genome sequencing reads generated for SCHI0154.S.1 (Australian Centre for Ecogenomics, St Lucia, Queensland, Australia) against 26 Pantoea spp. reference genomes (Appendix Table). We then analyzed those sequencing results against 48 publicly available P. stewartii genomes (Appendix Table). We carried out phylogenomic reconstruction of orthologous, biallelic single-nucleotide polymorphisms by using default SPANDx v4.0.3 (6) settings and PAUP* v4.0a.168 (7). We performed bootstrapping using 1,000 re-samples on the P. stewartii tree to assess clade confidence. We visualized trees in FigTree v1.4.0. We deposited the SCHI0154.S.1 assembly into GenBank (accession no. GCA_030144305.1).
Phylogenomic analysis revealed that SCHI0154.S.1 was most closely related to P. stewartii subsp. indologenes PANS 07–14 (Appendix Figure), which was isolated from a Verbena plant on an onion farm in Georgia, USA, in 2007 (8). SCHI0154.S.1 and PANS 07–14 differed by 1,015 single-nucleotide polymorphisms. In contrast, the only other genome-sequenced P. stewartii isolate from Australia, C10109_Jinnung (also subsp. indologenes), retrieved from a sick, captive western ground parrot (Pezoporus flaviventris), at Perth Zoo (Perth, WA, Australia) in 2021 (9), differed from SCHI0154.S.1 by 27,876 single-nucleotide polymorphisms.
One previous study reported P. stewartii associated with a human infection, with taxonomic assignment based on genetic similarity analysis of an unpublished 1,212-bp 16S ribosomal RNA amplicon reported to be 99.69% similar to P. stewartii strain 08BF11TN (GenBank accession KX146472.1) (1). To confirm this result, we repeated an ‘All genomes’ National Center for Biotechnology Information BLAST v2.15.0+ (https://blast.ncbi.nlm.nih.gov) analysis of the 1,451bp KX146472.1 sequence on August 16, 2024, using both the ‘Complete, Microbes’ and ‘Draft, Microbes’ databases. We restricted search parameters to ‘Pantoea (taxid:53335)’ and ‘megablast’. Our BLAST analysis found a closer match to P. agglomerans 33.1 (accession no. NZ_CP083809.1; 99.45% identity and 100% query coverage) than to P. stewartii RON18713 (accession no. NZ_CP116285.1; 98.07% identity and 100% query coverage) using the ‘Complete, Microbes’ genome database. Similarly, the same BLAST search using the ‘Draft, Microbes’ genome database identified a closer match to P. vagans 848 (accession no. JUQR01000382.1; 99.79% identity and 100% query coverage) and P. septica FF5 (accession no. CCAQ010000001.1; 99.72% identity and 100% query coverage) than to P. stewartii (best hit was strain RSA36 [accession no. LDSK01000027.1]; 98.00% identity and 100% coverage).
This unusual case confirms that the phytopathogen P. stewartii can cause life-threatening infections in humans. Although 1 published study described P. stewartii bacteraemia after a poststroke stent implantation in a patient from Spain (1), our repeat analysis suggested that this previously reported case was more likely caused by P. septica or P. agglomerans.
Because almost nothing is known about P. stewartii disease in humans or potential virulence factors of P. stewartii and its subspecies, this organism might be clinically underdiagnosed by current diagnostic methods, being misidentified as other more familiar Pantoea species. In support of this hypothesis, previously described P. agglomerans clinical isolates deposited into type culture collections have been reclassified as P. ananatis, Erwinia spp., or Enterobacter spp. on the basis of housekeeping gene sequencing (10). Further complicating matters, many taxonomic reassignments have occurred within the Erwiniaceae family in recent decades, making it challenging to track potential historical reports of P. stewartii human infection.
In our study, once the farmer’s infection was confirmed to be P. stewartii, we conducted a subsequent thorough clinical history to determine the likely source and mode of transmission. The patient reported no previous history of travel outside of Australia or recent injuries suggestive of dissemination from skin inoculation.
We noted just 1 previously report of P. stewartii in Australia, detected in a critically endangered native parrot that fell gravely ill in captivity (9), suggesting that birds may represent an underappreciated reservoir for P. stewartii subsp. indologenes global dissemination. However, in that case, the authors reported a link to parrot pellets commercially imported from the United States (9). It is therefore possible that P. stewartii was introduced into eastern Australia, and then to our patient’s macadamia farm, through a commercially imported agricultural product originating from the United States. In our study, although we could not determine the precise source of infection from the research conducted, the farmer’s clinical features (i.e., pneumonia with subsequent hematogenous dissemination), lack of clear inoculation source, and limited travel suggested the most likely mode of transmission to be inhalation from an environmental source on the farm.
Dr Subedi is an infectious diseases physician, a clinical microbiologist, and a PhD candidate at the University of Queensland Centre for Clinical Research. Her main research interests include evaluation and implementation of new and innovative methods of diagnosis of infection.
Acknowledgment
We wish to thank Rhys White for helpful discussions on Pantoea stewartii genomics.
References
- Cobo F, González A, Pérez-Carrasco V, García-Salcedo JA. Pantoea stewartii: A new pathogen as a cause of bacteremia? Enferm Infecc Microbiol Clin (Engl Ed). 2022;40:278–80. DOIPubMedGoogle Scholar
- Walterson AM, Stavrinides J. Pantoea: insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol Rev. 2015;39:968–84. DOIPubMedGoogle Scholar
- Mergaert J, Verdonck L, Kersters K. Transfer of Erwinia ananas (synonym, Erwinia uredovora) and Erwinia stewartii to the genus Pantoea emend. as Pantoea ananas (Serrano 1928) comb. nov. and Pantoea stewartii (Smith 1898) comb. nov., respectively, and description of Pantoea stewartii subsp. indologenes subsp. nov. Int J Syst Bacteriol. 1993;43:162–73. DOIGoogle Scholar
- Pataky J. Pest risk analysis: the risk of introducing Erwinia stewartii in maize seed [cited 2025 Apr 15]. https://worldseed.org/wp-content/uploads/2015/10/Erwinia_stewartii.pdf
- Bragard C, Dehnen-Schmutz K, Di Serio F, Gonthier P, Jacques MA, Jaques Miret JA, et al.; EFSA Panel on Plant Health (PLH). Risk assessment of the entry of Pantoea stewartii subsp. stewartii on maize seed imported by the EU from the USA. EFSA J. 2019;17:
e05851 .PubMedGoogle Scholar - Sarovich DS, Price EP. SPANDx: a genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets. BMC Res Notes. 2014;7:618. DOIPubMedGoogle Scholar
- Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4.0. Sunderland (MA): Sinauer Associates; 2003.
- Koirala S, Zhao M, Agarwal G, Gitaitis R, Stice S, Kvitko B, et al. Identification of two novel pathovars of Pantoea stewartii subsp. indologenes affecting Allium sp. and millets. Phytopathology. 2021;111:1509–19. DOIPubMedGoogle Scholar
- White RT, Taylor W, Klukowski N, Vaughan-Higgins R, Williams E, Petrovski S, et al. A discovery down under: decoding the draft genome sequence of Pantoea stewartii from Australia’s Critically Endangered western ground parrot/kyloring (Pezoporus flaviventris). Microb Genom. 2023;9:
001101 . DOIPubMedGoogle Scholar - Rezzonico F, Smits TH, Montesinos E, Frey JE, Duffy B. Genotypic comparison of Pantoea agglomerans plant and clinical strains. BMC Microbiol. 2009;9:204. DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: January 14, 2025
Table of Contents – Volume 31, Number 2—February 2025
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Shradha Subedi, University of Queensland Centre for Clinical Research, Bld 71/918 Rbwh Herston, Brisbane, QLD 4029, Australia
Top