Volume 31, Number 6—June 2025
Research Letter
Francisella tularensis Subspecies holarctica in Stranded Beluga Whales, Cook Inlet, Alaska, USA
Abstract
We report fatal tularemia in stranded beluga whales in Cook Inlet, Alaska, USA. Francisella tularensis was detected by nanopore metagenomics, confirmed by quantitative PCR and immunohistochemistry, and characterized as F. tularensis subspecies holarctica by multilocus sequence typing. Our findings should be considered when assessing biosecurity and marine mammal health in the North Pacific.
Francisella tularensis is a highly pathogenic gram-negative bacterium that infects a large range of animals and humans, primarily in the Northern Hemisphere, causing the clinical disease tularemia. Human disease manifests with influenza-like symptoms (lymphadenopathy, conjunctivitis, pneumonia, septicemia) and other specific symptoms corresponding to the route of exposure. Two subspecies, F. tularensis subsp. tularensis and holarctica, are known pathogens and can be acquired via multiple routes, including arthropod vector, cutaneous, ingestion, or inhalation (1).
F. tularensis was first documented in Alaska, USA, in 1938 (2) and has been isolated infrequently in ticks, lagomorphs, and rodents. Serologic studies have confirmed exposure in humans, avian species, terrestrial mammals, and polar bears in multiple areas of the state (2). In October 2023, tularemia was diagnosed in a pinniped in Washington, USA, when a biologist was infected during necropsy (3). The same fall, dead stranded beluga whales (Delphinapterus leucas) in Cook Inlet, Alaska, were found to have gross lesions consistent with tularemia. We report the results of an investigation of those deaths.
Necropsies were performed and tissues collected and stored following standard procedures. Samples for histopathology were fixed in 10% neutral buffered formalin (Table). We submitted varied tissues from 2 sufficiently fresh animals (no. 2023279: fetal spleen, mediastinal lymph node, spleen, blowhole swab, heart, liver; and no. 2023288: brain, liver, mammary gland, mediastinal lymph node, spleen) for aerobic culture and testing for known cetacean pathogens, including influenza and Erysipelothrix sp. by PCR, and for harmful algal bloom toxins by ELISA (Table). We analyzed blowhole swab, lung, mediastinal lymph node, and rectal swab samples from animal 2023279 by metagenomic sequencing. In brief, we extracted and amplified total nucleic acids (I.M. Claro et al., unpub. data, https://doi.org/10.12688/wellcomeopenres.17170.2) and sequenced cDNA and metagenomics libraries by SMART9N using an Oxford Nanopore Rapid PCR barcoding and MinION device (https://nanoporetech.com) (Table; Appendix Figure 1) (4). We classified sequence reads by using wf-metagenomics and wf-alignment in epi2melabs v.5.1.3 (Oxford Nanopore), mapped to the F. tularensis genome (GenBank accession no. NC_007880.1) by reference-based assembly using minimap2, and annotated using tbCon and ggplot in RStudio (Posit, https://posit.co) (Table). Subsequently, we tested lung and liver tissue from both animals for F. tularensis by immunohistochemistry and by culture and PCR using Centers for Disease Control and Prevention Laboratory Response Network proprietary protocols (Table). We then typed samples from positive animals by multilocus type sequencing of 6 genes (fabH, tpiA, sdhA, rpoA, groEL, and pgm) (5–7) and sequenced multiplexed amplicon libraries on the MiSeq platform (Illumina, https://www.illumina.com) (Table). We mapped amplicon sequence reads to reference genes from F. tularensis subsp. holarctica live vaccine strain, concatenated, and aligned with corresponding sequences from F. tularensis and other Francisella spp. to construct phylogenetic trees.
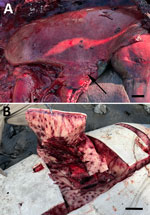
Both animals were pregnant adult females with markedly enlarged mediastinal lymph nodes, pleuritis, and pneumonia (Figure, panel A). One animal had severe multifocal random ecchymotic hemorrhage in the blubber (Figure, panel B). Histologic findings included necrosuppurative and histiocytic bronchopneumonia, lymphadenitis, and hepatitis (Appendix Figure 2, panels A–C). Immunohistochemistry demonstrated positive staining in areas of inflammation (Appendix Figure 2, panel D). Domoic acid and saxitoxin were not found, and PCRs and bacterial cultures yielded negative results or mixed organisms believed to be postmortem overgrowth (Appendix Table).
We identified the causative organism by using metagenomics. We mapped sequence reads from animal 2023279 by reference-based assembly and found those reads to be distributed at low read depth (2–21×; 1,181 sequence reads; N50 = 275 nt, quality score = 9) across the 1.89-Mbp F. tularensis genome. We detected F. tularensis DNA in all samples by quantitative PCR with cycle threshold values <25. By multilocus sequence typing, we identified a concatenated sequence of 4,107 bp as F. tularensis subsp. holarctica. Phylogenetic analysis placed this strain in a clade identical to the 2023 pinniped case from Washington, as well as other isolates from the Northern Hemisphere (Appendix Figure 3).
Although Cook Inlet belugas are known to be susceptible to a variety of bacterial pathogens (10), F. tularensis has not been previously detected in this population, or in other cetaceans. The pattern of pathology represents the pulmonary form of tularemia, and the route of exposure was likely inhalation of contaminated water. F. tularensis is primarily a disease associated with freshwater, but the brackish nature of Cook Inlet and nearshore residence of belugas expose them to potentially contaminated freshwater runoff as well as to other reservoirs typically associated with freshwater (e.g., aquatic rodents, mosquito larvae) (1,2). The cause of the infections in a previously unreported host is unknown; however, host factors such as immunosuppression or environmental changes, such as increased runoff, could be considered.
One human case of tularemia was reported in Cook Inlet’s largest adjacent city in 2023 (https://epi.alaska.gov/bulletins/docs/b2024_14.pdf); however, the circumstances of exposure were not reported. The propensity of whales to travel long distances could further disseminate this pathogen, increasing exposure to humans and wildlife. Our findings highlight a new risk to persons working in the marine environment and should be considered when assessing biosecurity and marine mammal health in the North Pacific.
Ms. Rouse is a necropsy biologist at Alaska Veterinary Pathology Services, Eagle River, Alaska. Her main interests are wildlife health and disease ecology.
Acknowledgments
We thank the Alaska Marine Mammal Stranding network, Melanie Iverson, and the National Marine Fisheries Service Protected Species Division for assistance with field sampling, and Hailei Nystuen, Soren George-Nichol, and Bryce Inman for assistance with molecular analysis.
All samples were collected under National Oceanic and Atmospheric Administration permit no. 24359. Diagnostics not related to these findings were run at University of California Davis and University of Georgia Veterinary Diagnostic lab (aerobic culture), Tufts Puyear lab (viral PCRs), and WARRN West (harmful algal bloom toxin testing). We deposited sequences from this study into GenBank (accession nos. PQ724310–21) and the National Center for Biotechnology Information Sequence Read Archive (accession nos. SRR31713860 and SRR31713861) (Appendix).
Funding for stranding response, molecular analyses, and manuscript preparation was provided by the John H. Prescott grant program (grant nos. NA22NMF4390247 and NA23NMF4390303) and National Marine Fisheries Service contract no. 1333MF22PNFFS0225. Funding for pathogen genomics analysis was provided by the National Institutes of Health National Institute of General Medical Sciences IDeA Networks of Biomedical Research Excellence grant no. P20GM103395 (Alaska INBRE), US Department of Agriculture American Rescue Plan SPASAK project subaward to U.A.A. (Animal Research Service grant no. AP23OA000000C014), and the National Institutes of Health National Institute of Allergy and Infectious Diseases Centers of Excellence for Influenza Research and Response (contract no. 75N93021C00014).
References
- Hennebique A, Boisset S, Maurin M. Tularemia as a waterborne disease: a review. Emerg Microbes Infect. 2019;8:1027–42. DOIPubMedGoogle Scholar
- Smith MM, Van Hemert C, Atwood TC, Sinnett DR, Hupp JW, Meixell BW, et al. A serologic survey of Francisella tularensis exposure in wildlife on the Arctic Coastal Plain of Alaska, USA. J Wildl Dis. 2022;58:746–55. DOIPubMedGoogle Scholar
- Inouye W, Oltean HN, McMillan M, Schnitzler H, Lipton B, Peterson JM, et al. Notes from the field: tularemia associated with harbor seal necropsy—Kitsap County, Washington, October 2023. MMWR Morb Mortal Wkly Rep. 2024;73:731–2. DOIPubMedGoogle Scholar
- Buttler J, Drown DM. Accuracy and completeness of long read metagenomic assemblies. Microorganisms. 2022;11:96. DOIPubMedGoogle Scholar
- Ahlinder J, Öhrman C, Svensson K, Lindgren P, Johansson A, Forsman M, et al. Increased knowledge of Francisella genus diversity highlights the benefits of optimised DNA-based assays. BMC Microbiol. 2012;12:220. DOIPubMedGoogle Scholar
- Mikalsen J, Olsen AB, Tengs T, Colquhoun DJ. Francisella philomiragia subsp. noatunensis subsp. nov., isolated from farmed Atlantic cod (Gadus morhua L.). Int J Syst Evol Microbiol. 2007;57:1960–5. DOIPubMedGoogle Scholar
- Brett M, Doppalapudi A, Respicio-Kingry LB, Myers D, Husband B, Pollard K, et al. Francisella novicida bacteremia after a near-drowning accident. J Clin Microbiol. 2012;50:2826–9. DOIPubMedGoogle Scholar
- Pal N, Bender JS, Opriessnig T. Rapid detection and differentiation of Erysipelothrix spp. by a novel multiplex real-time PCR assay. J Appl Microbiol. 2010;108:1083–93. DOIPubMedGoogle Scholar
- Puryear WB, Keogh M, Hill N, Moxley J, Josephson E, Davis KR, et al. Prevalence of influenza A virus in live-captured North Atlantic gray seals: a possible wild reservoir. Emerg Microbes Infect. 2016;5:
e81 . DOIPubMedGoogle Scholar - Rouse NM, Burek-Huntington K. Cook Inlet beluga whale Delphinapterus leucas with valvular endocarditis, encephalitis, rhabdomyolysis, heavy parasitism and fungal dermatitis. Dis Aquat Organ. 2023;155:1–6. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleOriginal Publication Date: May 14, 2025
Table of Contents – Volume 31, Number 6—June 2025
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Natalie Rouse, University of Alaska Anchorage, 3211 Providence Dr, Anchorage, AK 99508, USA
Top