Volume 9, Number 4—April 2003
Dispatch
Automated Ribotyping and Pulsed-Field Gel Electrophoresis for Rapid Identification of Multidrug-Resistant Salmonella Serotype Newport
Cite This Article
Citation for Media
Abstract
In a series of 116 Salmonella enterica Newport isolates that included 64 multidrug-resistant (MDR) isolates, automated ribotyping and pulsed-field gel electrophoresis (PFGE) discriminated MDR S. Newport with a sensitivity of 100% and 98% and specificity of 76% and 89%, respectively. Clustering of PFGE patterns (but not ribotyping) linked human and bovine cases. Automated ribotyping rapidly identified the MDR strain, and PFGE detected associations that aided epidemiologic investigations.
An eight-drug resistant strain of Salmonella enterica serotype Newport (multidrug-resistant [MDR] S. Newport) characterized by resistance to at least ampicillin, cephalothin, chloramphenicol, clavulanic acid, streptomycin, sulfamethoxazole, tetracycline, and ceftriaxone (intermediate to complete) recently identified by the National Antimicrobial Resistance Monitoring System (NARMS), Centers for Disease Control and Prevention (CDC) (1), is an example of the emergent problem of antibiotic resistance. However, only a few state laboratories routinely perform susceptibility testing on enteric organisms, particularly in a format comparable to that employed by NARMS. Rapid identification of drug resistance is critical for preventing and treating diseases and for epidemiologic analysis (2).
Pulsed-field gel electrophoresis (PFGE) testing is more common at state public health laboratories; 50 such laboratories participate in CDC’s PFGE surveillance program, PulseNet. In PulseNet, participants submit PFGE data on E. coli O157:H7, Salmonella spp., Shigella spp., and Listeria monocytogenes to a national database (3). Because PulseNet laboratories follow a standardized protocol, PFGE patterns can be compared reliably within the network and associations made promptly among isolates that may be few in number and widely separated geographically.
However, the clonal nature or inherent genetic variability of bacteria may limit the ability of PFGE either to link isolates or to detect relatedness among a set of isolates from a single outbreak (4). In some organisms, such as Shigella spp. and Campylobacter spp., PFGE patterns vary considerably, and differences in banding patterns can occur among isolates that are epidemiologically linked (5). In other organisms, such as Salmonella serotype Enteritidis, variability of PFGE patterns is limited, and banding patterns can be indistinguishable among isolates that are not epidemiologically linked. Often, supplemental methods are needed to detect an association among isolates because of their apparent clonal characteristics (6). Therefore, PFGE databases must contain patterns from a sufficient number of isolates of a species representative of circulating strains to enable accurate interpretation of relatedness.
In Massachusetts, S. Newport isolates analyzed by PFGE have shown a high degree of variability. Ribotyping has been reported to be less discriminatory than PFGE (7) but can provide information that identifies real associations undetected solely by PFGE analysis, particularly when epidemiologic data are limited (8). We evaluated the usefulness of PFGE and automated ribotyping, independently and together, to monitor and characterize the MDR strain of S. Newport in Massachusetts.
Specimens of Salmonella spp. submitted to the State Laboratory Institute as pure cultures or isolates from fresh stool samples submitted in Meridian Para-Pak C & S medium (Meridian Bioscience, Inc., Cincinnati, OH) were serotyped by the Kaufmann-White scheme, according to CDC protocols (9). After an increase in the incidence of S. Newport , the Massachusetts Department of Public Health initiated enhanced surveillance for this bacterium. In December 2000, we posted patterns MA-JJP0034 and MA-JJP0070 on the PulseNet listserv and asked other states to determine detection of S. Newport (CDC PulseNet patterns JJP.X01.0014 and JJP.X01.0181, respectively). In response, Oklahoma, Minnesota, Maine, and Vermont posted similar or indistinguishable isolates. In January 2001, S. Newport was isolated from a stool specimen from an employee at a Massachusetts farm that also reported diarrheal illness in cows. Investigators from the Massachusetts Department of Public Health and the Massachusetts Department of Food and Agriculture, Bureau of Animal Health, with assistance from CDC, visited farms and auction houses to obtain stool specimens from cows and calves exhibiting diarrheal illness. Isolates identified as S. Newport were tested by PFGE and automated ribotyping, and for antimicrobial resistance.
All S. Newport isolates were tested for resistance to amikacin, ampicillin, amoxicillin/clavulanic acid, apramycin, cefoxitin, ceftiofur, ceftriaxone, cephalothin, chloramphenicol, ciprofloxacin, gentamicin, kanamycin, nalidixic acid, streptomycin, sulfamethoxazole, tetracycline, and trimethoprim/sulphamethoxazole by using the Trek Diagnostics Sensititre CMV1UIL (Trek Diagnostic Systems, Inc., Cleveland, OH) plate. Isolates categorized as MDR S. Newport were resistant to at least ampicillin, cephalothin, chloramphenicol, clavulanic acid, streptomycin, sulfamethoxazole, and tetracycline, and demonstrated complete or intermediate resistance to ceftriaxone. An E. coli strain, ATTC 25922, was used as a control organism during each susceptibility test run.
PFGE was performed on all S. Newport isolates, according to previously described methods (10). Automated ribotyping was performed by using the Riboprinter Microbial Characterization System (DuPont Qualicon, Wilmington, DE), according to the manufacturer’s protocol. Briefly, a sweep of pure S. Newport cells from each test isolate was suspended in a 12- x 75-mm Falcon tube in sample buffer to 20% transmittance in a Vitek Colorimeter (bioMérieux, Durhan, NC). Thirty microliters of each suspension was transferred to one well in an eight-well sample carrier and heat treated to 80°C for 30 min; 5 µL of lysis buffers A and B was then added. The sample carrier was then placed in the Riboprinter for automated processing, which included the following steps: DNA extraction, PvuII restriction and electrophoresis through a 1% agarose gel, DNA transfer, and hybridization to a nylon membrane. The DNA was probed with a sulfonated 1,020-bp E. coli rrn, which was detected with alkaline phosphatase–labeled anti-sulfonated anti-DNA antibodies. Ribotype images were captured by a charge-coupled device camera and compared for similarity to images in a PvuII database of 272 Salmonella patterns of 138 serotypes (11).
PFGE patterns were analyzed with Molecular Analyst Software Version 1.11 (Bio-Rad Laboratories, Hercules, CA) by using the Dice coefficient with a band position tolerance of 1.5%. This method considers only the presence or absence of a band. All S. Newport patterns were compared with pattern MA-JJP0034, the first pattern of MDR S. Newport that appeared in Massachusetts. Patterns with a similarity coefficient of >0.85 relative to pattern MA-JJP0034 were designated in the analysis as MDR S. Newport by PFGE (D. Boxrud, pers. comm.).
Ribotype patterns were analyzed by the Riboprinter software, using the Pearson coefficient against a database of unique patterns of Salmonella serotypes. In this study, the S. Newport ribotypes with a similarity >0.85. were included in the MDR ribogroup. Sensitivity and specificity of the PFGE and ribotyping methods to identify MDR S. Newport were calculated and compared to the test results obtained by serotyping and antibiotic sensitivity testing for the 130 S. Newport isolates (12).
One hundred sixteen S. Newport isolates from 300 human specimens and 14 S. Newport isolates from 50 bovine specimens were identified by serotyping at the State Laboratory Institute during the study period; 64 isolates were identified as MDR S. Newport (50 [43%] of 116 human isolates and all 14 bovine isolates). In addition to the eight-drug resistance pattern of the MDR isolates, 11 of 50 human isolates and 11 of 14 bovine isolates were resistant to kanamycin.
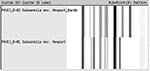
Two ribotype patterns, D-81 and D-82, designated as S. Newport in the Riboprinter database, were associated with the 130 S. Newport serotyped isolates (Figure 1). The 64 MDR S. Newport isolates, 50 human isolates, and 14 bovine isolates, matched only to ribotype pattern D-81. Sixteen of 66 pansusceptible S. Newport human isolates also were identified as D-81 ribotype pattern. The remaining 50 pansusceptible S. Newport isolates were identified as D-82 ribotype pattern (Table). Using pattern D-81, ribotyping had a sensitivity of 100% and a specificity of 76% in identifying MDR S. Newport among S. Newport isolates.
Among the 130 human and bovine S. Newport isolates were 58 PFGE patterns of XbaI digests (Figure 2). Eight PFGE patterns were associated with 64 MDR S. Newport isolates (Figure 3). Two of the 66 pansusceptible isolates smeared by PFGE were not interpretable, leaving 64 isolates for PFGE analysis. The most common PFGE pattern was MA-JJP0034, which appeared in 41 (64%) of 64 of the MDR S. Newport isolates. MA-JJP00015 was the most common pattern among the pansusceptible isolates, being found in 7 (11%) of 64 isolates. When the S. Newport isolates were analyzed against PFGE pattern 34, all MDR S. Newport isolates and 7 of 64 pansusceptible S. Newport isolates had a similarity coefficient of >0.85. Thus, PFGE had a sensitivity of 98% and a specificity of 89% when this pattern was used to screen this set of isolates.
The 64 MDR S. Newport, among a sample of 130 S. Newport isolates, were correctly identified by comparing the sensitivity and specificity of PFGE and automated ribotyping to conventional serotyping and antibiotic susceptibility test results. The sensitivity of each method was >98%. The specificity of PFGE and automated ribotyping was 89% and 76%, respectively, with no false-negative results, and 11% and 24%, respectively, of S. Newport isolates misidentified (false positive) as the MDR strain.
Automated ribotyping was a rapid means of subtyping S. Newport and distinguishing the MDR strain from non-MDR strains by using the Riboprinter PvuII database. This capability is important because most often Salmonella isolates are submitted to state laboratories and characterized to the O antigen level, not the species level. Further, serotyping performed at a state public health laboratory to define the Salmonella species requires a minimum of 2 days, and for biphasic Salmonella species, such as S. Newport a minimum of 4 days (9). PFGE was useful in distinguishing associations within the isolates of S. Newport, which allowed for identification of potential epidemiologically related events, such as the PFGE pattern of MDR S. Newport common among a subset of bovine and human isolates. After a pure culture isolate is obtained, turnaround times for PFGE and ribotyping testing are approximately 24 h and 8 h, respectively; however, serotype identification often is required for interpretation of PFGE test results, depending on the organism, which can lengthen turnaround for final results by 1 or more days.
Both automated ribotyping and PFGE rely on existing databases of images from individual isolates to characterize DNA fingerprints. As data from more strains are added to databases, characterizing similarities or differences between isolates becomes easier. Because the fingerprint database of the PulseNet program is extensive and PFGE is more discriminatory that ribotyping, PFGE provides a more robust tool in characterizing the development of emerging pathogens.
These data suggest that PFGE and ribotyping can be used together to provide rapid identification of the MDR strain of S. Newport. These methods offer important capabilities for laboratory-based surveillance for public health purposes; however, automated ribotyping is very costly and should be used in a public health laboratory only if justified by appropriate cost-benefit analysis. Furthermore, these data support the need for strengthening public health laboratory infrastructure to facilitate early detection of infectious disease risks.
Dr. Fontana is director of the Pulsed Field Gel Electrophoresis and Antimicrobial Resistance Laboratory at the Massachusetts Department of Public Health/State Laboratory Institute. With the support of the State Laboratory Institute and epidemiologists in the Division of Communicable Disease, Dr. Fontana established the Antimicrobial Resistance Surveillance Laboratory in 1999, which performs susceptibility testing on invasive Streptococcus pneumoniae, vancomycin resistant enterococci, Salmonella spp., Shigella spp., and Escherichia coli O157:H7.
References
- Joyce K, Mshar P, Sexton J, Goddard A, Fedorka-Cray P, Rossiter S, ; NARMS Working Group. Emergence of a multidrug-resistance strain of Salmonella Serotype Newport in the United States: NARMS 1997–1999. In: Program and abstracts of International Conference on Emerging Infectious Diseases Atlanta, Georgia, July 2000. Washington: American Society for Microbiology; 2000.
- Hughes JM, Tenover FC. Approaches to limiting emergence of antimicrobial resistance in bacteria in human populations. Clin Infect Dis. 1997;24(Suppl 1):S131–5.PubMedGoogle Scholar
- Swaminathan B, Barrett TJ, Hunter SB, Tauxe RV. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg Infect Dis. 2001;7:382–9.PubMedGoogle Scholar
- Tenover FC, Arbeit RD, Goering RV. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. Molecular Typing Working Group of the Society for Healthcare Epidemiology of America. Infect Control Hosp Epidemiol. 1997;18:426–39. DOIPubMedGoogle Scholar
- Sobel J, Cameron DN, Ismail J, Strockbine N, Williams M, Diaz PS, A prolonged outbreak of Shigella sonnei infections in traditionally observant Jewish communities in North America caused by a molecularly distinct bacterial subtype. J Infect Dis. 1998;177:1405–9. DOIPubMedGoogle Scholar
- Helmuth R, Schroeter A. Molecular typing methods for S. enteritidis. Int J Food Microbiol. 1994;21:69–77. DOIPubMedGoogle Scholar
- Liebana E, Guns D, Garcia-Migura L, Woodward MJ, Clifton-Hadley FA, Davies RH. Molecular typing of Salmonella serotypes prevalent in animals in England: assessment of methodology. J Clin Microbiol. 2001;39:3609–16. DOIPubMedGoogle Scholar
- Quale JM, Landman D, Flores C, Ravishankar J. Comparison of automated ribotyping to pulsed-field gel electrophoresis from genetic fingerprinting of Streptococcus pneumoniae. J Clin Microbiol. 2001;39:4175–7. DOIPubMedGoogle Scholar
- Brenner F, McWhorten-Murlin AC. Identification and serotyping of Salmonella. Atlanta (GA): Centers for Disease Control and Prevention, National Salmonella Reference Laboratory; 1998.
- Ribot E, Wierzba R, Angulo F, Barrett T. Salmonella enterica serotype Typhimurium DT104 isolated from humans, United States, 1985, 1990, and 1995. Emerg Infect Dis. 2002;8:387–91. DOIPubMedGoogle Scholar
- Graves L, Holland B, Swaminathan B, DeMartino M, Moyer N, Fontant J, Assessment of the utility of an automated ribotyping network for the National Salmonella Surveillance System in the United States. In: Program and abstracts of the American Society for Microbiology general meeting, Salt Lake City, Utah, May 19–23, 2002. Washington: American Society for Microbiology; 2002.
- Sackett DL, Haynes RB, Tugwell P. The interpretation of diagnostic data. In: Sackett, DL, Haynes RB, Tugwell P, editors. Clinical epidemiology: a basic science for clinical medicine. Boston: Little, Brown; 1985. p.59–138.
Figures
Table
Cite This ArticleTable of Contents – Volume 9, Number 4—April 2003
| EID Search Options |
|---|
|
|
|
|
|
|


Please use the form below to submit correspondence to the authors or contact them at the following address:
John Fontana, Massachusetts Department of Public Health, State Laboratory Institute, 305 South Street, Jamaica Plain, MA 020130, USA; fax: 617-983-6618
Top