Volume 16, Number 7—July 2010
Dispatch
Detection of Lassa Virus, Mali
Figure 2
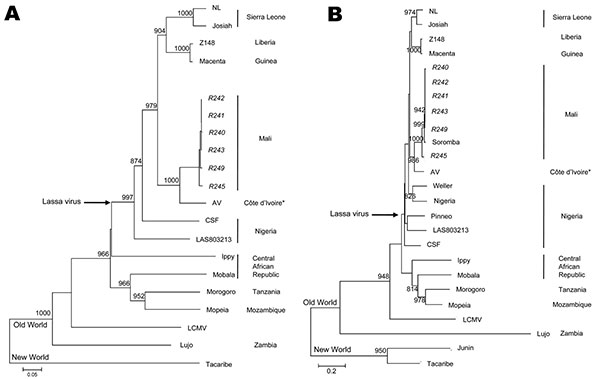
Figure 2. Phylogenetic analysis of Lassa virus conducted on A) a 754-bp fragment of the polymerase gene (large genomic segment nucleotide positions 3427–4180) and B) a 771-bp fragment of the glycoprotein precursor (small genomic segment nucleotide positions 2526–3296). The fragments were amplified from infected rodent tissues with sequence analysis accomplished with PHYLIP version 3.69 software (http://evolution.genetics.washington.edu/phylip.html) by using the neighbor-joining method with 1,000 replicates of bootstrap. Sequences were compared with the following arenavirus sequences (small and large segment GenBank accession nos.): Tacaribe (NC_004293, NC_004292), Lujo (NC_012776, NC_012777), Lymphocytic choriomeningitis virus (LCMV, strain Armstrong, AY847350,J04331), Ippy (NC_007905, NC_007906), Mobala (NC_007903, NC_007904), Morogoro (NC_013057, NC_013058), Mopeia (NC_006575, NC_006574), and Lassa virus strains 803213 (AF803213, AY693640), CSF (AF333969, AY179174), NL (AY179173, AY179172), Josiah (AY628203, U63094), Z148 (AY628205, AY628204), Macenta (AY628201, AY628200), and AV (AF246121, AY179171). Small segment analysis also included Lassa virus strains LP (AF18185), Weller (AY628206), Nigeria (M36544), the sequence generated from the imported case of Lassa fever from Mali (FJ824031), and Junin virus (NC_005081). Bootstrap values >800 are shown. *Whether the AV strain of Lassa virus originated from Côte d’Ivoire, Burkina Faso, or Ghana is not clear. Italicized names represent sequences generated as part of these studies. Scale bars indicate 5% (A) or 20% (B) nucleotide divergence.