Volume 8, Number 1—January 2002
Research
Genetic Characterization of the M RNA Segment of Crimean Congo Hemorrhagic Fever Virus Strains, China
Figure 2
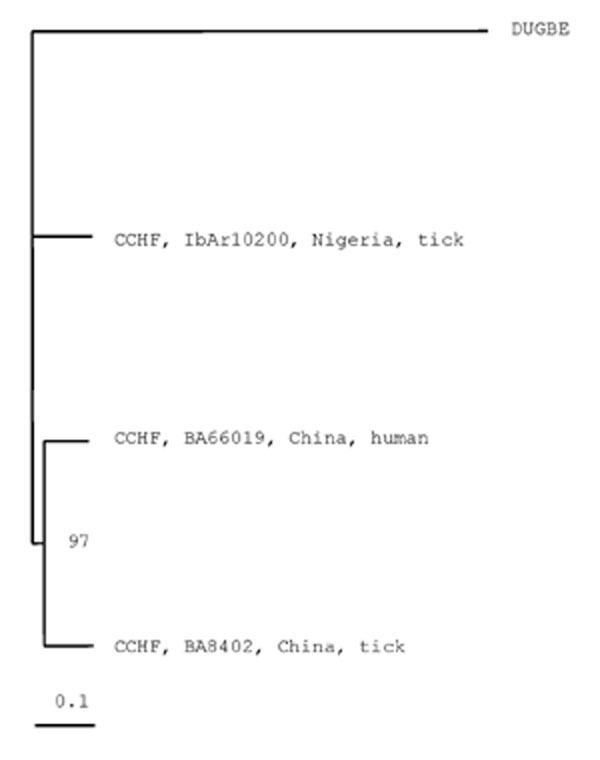
Figure 2. Phylogenetic tree based on 4,722 nt of the medium (M) RNA segment, including the two Chinese Crimean Congo hemorrhagic fever virus (CCHFV) strains BA66019 and BA8402 and the CCHFV strain IbAr10200, GenBank accession number U39455. Dugbe virus (DUGV) strain ArD44313, GenBank accession number M94133, was used as outgroup. Horizontal distances are proportional to nucleotide difference; vertical distances are for graphic display only. Bootstrap support (in %) is indicated at the respective branch.
Page created: July 14, 2010
Page updated: July 14, 2010
Page reviewed: July 14, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.