Volume 9, Number 8—August 2003
Research
Molecular Characterization of a Non–Babesia divergens Organism Causing Zoonotic Babesiosis in Europe
Figure 1
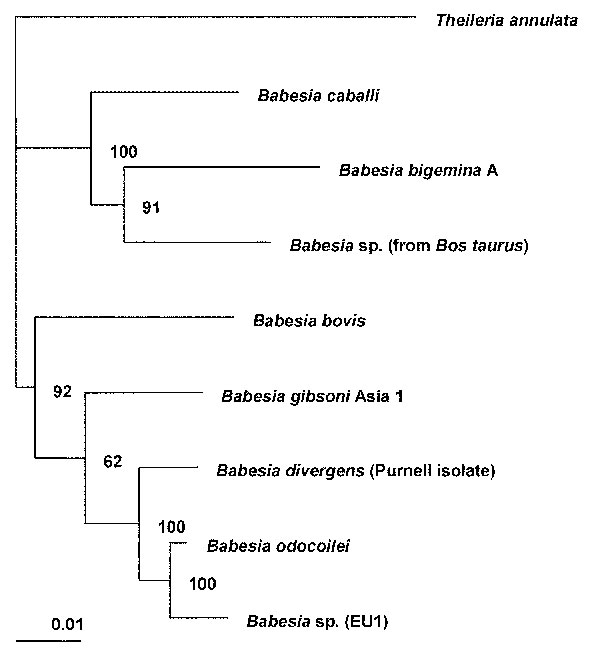
Figure 1. Phylogenetic tree for the complete 18S rRNA gene from selected Babesia spp. The tree was computed by using the quarter puzzling maximum likelihood method of the TREE-PUZZLE program and was oriented by using Theileria annulata as the outgroup. Numbers at the nodes indicate the quartet puzzling support for each internal branch. Scale bar indicates an evolutionary distance of 0.01 nucleotides per position in the sequence. Vertical distances are for clarity only. The GenBank accession numbers for the sequences used in the analysis are as follows: Babesia bigemina A, X59604; B. bovis, L19077; B. caballi, Z15104; B. divergens (Purnell isolate [12]), AY046576; B. gibsoni (genotype Asia 1), AF175300; B. odocoilei (Brushy Creek and Engeling isolates [14]), AY046577; Babesia sp. (isolated from Bos taurus), U09834; EU1 (the etiologic agent of infection in the two cases described here), AY046575; and Theileria annulata, M64243.
1The issue of what constitutes a new or newly described species requires periodic reevaluation as the techniques for characterizing microbes improve. Although the advent of molecular biology/phylogeny has made it possible to characterize organisms more precisely, the issue of how large a genetic difference in a particular gene(s) constitutes a new or different species is controversial. Bacterial taxonomy is a much more active and advanced field (26,27) than that for characterizing protozoa, in part because of the increasingly large numbers of bacteria being proposed as new species and the challenges posed by such possibilities as genetic rearrangements. In the recommendations published in 2002 by an ad hoc committee that reevaluated the species definition for bacteria (26), scientists were encouraged to use the “Candidatus” concept (i.e., to propose candidates for newly described bacterial species) for organisms that had been well-characterized, including the sequencing of the small subunit RNA gene, but had not yet been cultured. As noted above, we have referred to the protozoan we characterized as EU1. We have not claimed it as a newly identified species, despite having complete, identical, and novel 18S rRNA sequences for the organism from two patients, who were separated in time and space, and having done the sequence analysis for the two cases in different countries. However, although the Candidatus concept per se does not formally exist now for protozoa, on the basis of the precedent from the field of bacterial taxonomy, we propose that EU1 be considered a candidate species. If additional evidence supports the conclusion that the organism indeed constitutes a newly described species of the Babesia genus, we would favor the name Babesia venatorum, which now does not constitute an official name. We chose this name because the patients whose cases we reported were avocational hunters; “venator” is the Latin word for “hunter” (“venatorum,” the plural genitive case, means “of the hunters”).