Volume 11, Number 12—December 2005
Research
Intergenogroup Recombination in Sapoviruses
Figure 2
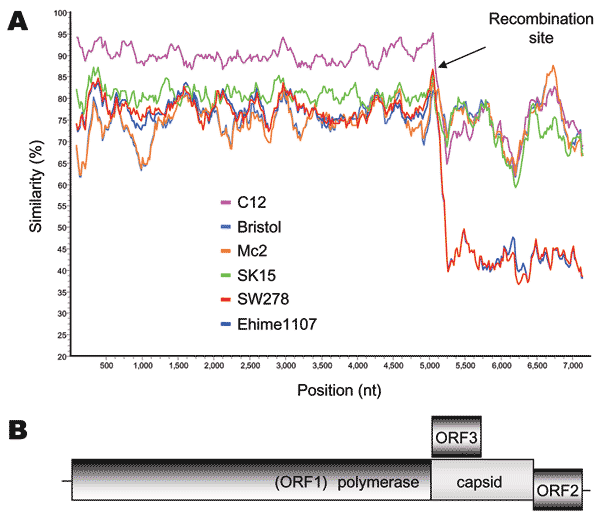
Figure 2. A) SimPlot analysis of 7 sapovirus (SaV) complete genome sequences. The Mc10 genome sequence was compared to C12, Bristol, Mc2, SK15, SW278, and Ehime1107 by using a window size of 100 bp with an increment of 20 bp. All gaps were removed. The recombination site is suspected to be located between the polymerase and capsid gene, as shown by the arrows. B) Genomic organization of the SaV SW278 and Ehime1107 strains.
Page created: February 02, 2012
Page updated: February 02, 2012
Page reviewed: February 02, 2012
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.