Volume 11, Number 9—September 2005
Dispatch
Divergent HIV and Simian Immunodeficiency Virus Surveillance, Zaire
Figure
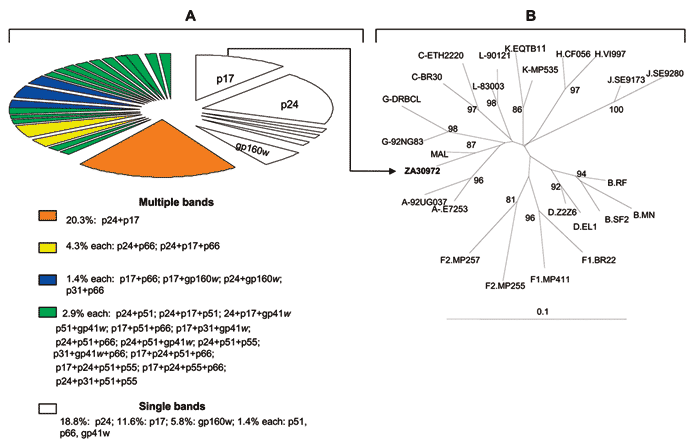
Figure. . A) Distribution of HIV-1 Western blot–indeterminate patterns among 69 serum specimens from Kinshasa, Zaire, reactive by enzyme immunoassay. B) Phylogenetic classification of HIV-1 protease sequence ZA30972 (GenBank accession no. AY562558) isolated from the p17 gag-reactive serum. The phylogenetic tree was generated by the neighbor-joining method with the nucleotide distance calculated by Kimura 2-parameter method (12), included in the GeneStudio package (http://www.genestudio.com). Reference sequences were obtained from the Los Alamos database (http://hiv-web.lanl.gov/MAP/hivmap.html). The position of the outgroup (Simian immunodeficiency virus [SIV]cpz) is not shown. Values on branches represent the percentage of 100 bootstrap replicates. The scale bar indicates an evolutionary distance of 0.10 nucleotides per position in the sequence.
References
- Sharp PM, Bailes E, Chaudhuri RR, Rodenburg CM, Santiago MO, Hahn BH. The origins of acquired immune deficiency syndrome viruses: where and when? Philos Trans R Soc Lond B Biol Sci. 2001;356:867–76. DOIPubMedGoogle Scholar
- Apetrei C, Metzger MJ, Richardsin D, Ling B, Telfer PT, Reed P, Detection and partial characterization simian immunodeficiency virus SIVmn strains from bush meat samples from rural Sierra Leone. J Virol. 2005;79:2631–6. DOIPubMedGoogle Scholar
- Peeters M, Courgnaud V, Abela B, Auzel P, Pourrut X, Bibollet-Ruche F, Risk to human health from a plethora of simian immunodeficiency viruses in primate bushmeat. Emerg Infect Dis. 2002;8:451–7.PubMedGoogle Scholar
- Souquiere S, Bibollet-Ruche F, Robertson DL, Makuwa M, Apetrei C, Onanga R, Wild Mandrillus sphinx are carriers of two types of lentivirus. J Virol. 2001;75:7086–96. DOIPubMedGoogle Scholar
- Kalish ML, Ndongmo CB, Wolfe ND, Fonjungo P, Alemnji G, Zeh C, Evidence for continued exposure to and possible infection of humans with SIV. In: Tenth international workshop on HIV dynamics and evolution; 2003 Apr 13–16; Lake Arrowhead, California; 2003.
- Kleinman S, Fitzpatrick L, Secord K, Wilke D. Follow-up testing and notification of anti-HIV Western blot atypical (indeterminant) donors. Transfusion. 1988;28:280–2. DOIPubMedGoogle Scholar
- Povolotsky J, Gold JW, Chein N, Baron P, Armstrong D. Differences in human immunodeficiency virus type 1 (HIV-1) anti-p24 reactivities in serum of HIV-1–infected and uninfected subjects: analysis of indeterminate Western blot reactions. J Infect Dis. 1991;163:247–51. DOIPubMedGoogle Scholar
- Delaporte E, Peeters M, Simon F, Dupont A, Schrijvers D, Kerouedan D, Interpretation of antibodies reacting solely with human retroviral core proteins in western equatorial Africa. AIDS. 1989;3:179–82. DOIPubMedGoogle Scholar
- Huet T, Dazza MC, Brun-Vezinet F, Roelants GE, Wain-Hobson S. A highly defective HIV-1 strain isolated from a healthy Gabonese individual presenting an atypical Western blot. AIDS. 1989;3:707–15. DOIPubMedGoogle Scholar
- Kalish ML, Robbins KE, Pieniazek D, Schaefer A, Nzilambi N, Quinn TC, Recombinant viruses and early global HIV-1 epidemic. Emerg Infect Dis. 2004;10:1227–34.PubMedGoogle Scholar
- Felsenstein J. PHYLIP-phylogeny interference package (version 3.2). Cladistics. 1989;5:164–6.
- Yang C, Dash BC, Simon F, van der Groen G, Pieniazek D, Gao F, Detection of diverse variants of human immunodeficiency virus–1 groups M, N, and O and simian immunodeficiency viruses from chimpanzees by using generic pol and env primer pairs. J Infect Dis. 2000;181:1791–5. DOIPubMedGoogle Scholar
- Pieniazek D, Ellenberger D, Janini LM, Ramos AC, Nkengasong J, Sassan-Morokro M, Predominance of human immunodeficiency virus type 2 subtype B in Abidjan, Ivory Coast. AIDS Res Hum Retroviruses. 1999;15:603–8. DOIPubMedGoogle Scholar
- Masciotra S, Yang C, Pieniazek D, Thomas C, Owen SM, McClure HM, Detection of simian immunodeficiency virus in diverse species and of human immunodeficiency virus type 2 by using consensus primers within the pol region. J Clin Microbiol. 2002;40:3167–71. DOIPubMedGoogle Scholar
- Alizon M, Montagnier L. Genetic variability in human immunodeficiency viruses. Ann N Y Acad Sci. 1987;511:376–84. DOIPubMedGoogle Scholar