Identical Genotype B3 Sequences from Measles Patients in 4 Countries, 2005
Jennifer Rota*

, Luis Lowe*, Paul A. Rota*, William Bellini*, Susan Redd*, Gustavo Dayan*, Rob van Binnendijk†, Susan Hahné†, Graham Tipples‡, Jeannette Macey§, Rita Espinoza¶, Drew Posey*, Andrew Plummer*, John Bateman*, José Gudiño#, Edith Cruz-Ramirez#, Irma Lopez-Martinez#, Luis Anaya-Lopez**, Teneg Holy Akwar††, Scott Giffin††, Verónica Carrión‡‡, Jan Felix Drexler‡‡, Andrea Vicari‡‡, Christina Tan§§, Bruce Wolf§§, Katherine Wytovich§§, Peter Borus¶¶, Francis Mbugua¶¶, Paul Chege¶¶, Janeth Kombich¶¶, Chantal Akoua-Koffi##, Sheilagh Smit***, Henry Bukenya†††, Josephine Bwogi†††, Frederick Ndhoga Baliraine†††, Jacques Kremer‡‡‡, Claude Muller‡‡‡, and Sabine Santibanez§§§
Author affiliations: *Centers for Disease Control and Prevention, Atlanta, Georgia, USA; †National Institute for Public Health and the Environment, Biltoven, the Netherlands; ‡Public Health Agency of Canada, Winnipeg, Manitoba, Canada; §Public Health Agency of Canada, Tunney's Pasture, Ottawa, Ontario, Canada; ¶Texas Department of State Health Services, Austin, Texas, USA; #Instituto de Diagnóstico y Referencia Epidemiológicos, Mexico City, Mexico; **Dirección General de Epidemiología de la Secretaria de Salud, Mexico City, Mexico; ††New Brunswick Department of Health, Fredericton, New Brunswick, Canada; ‡‡Pan American Health Organization, Washington, DC, USA; §§New Jersey Department of Health and Senior Services, Trenton, New Jersey, USA; ¶¶Kenya Medical Research Institute, Nairobi, Kenya; ##Institut Pasteur, Abidjan, Côte d'Ivoire; ***National Institute of Communicable Disease, Johannesburg, South Africa; †††Uganda Virus Research Institute, Entebbe, Uganda; ‡‡‡Laboratoire National de Santé, Luxembourg; §§§Robert Koch-Institute, Berlin, Germany
Main Article
Figure
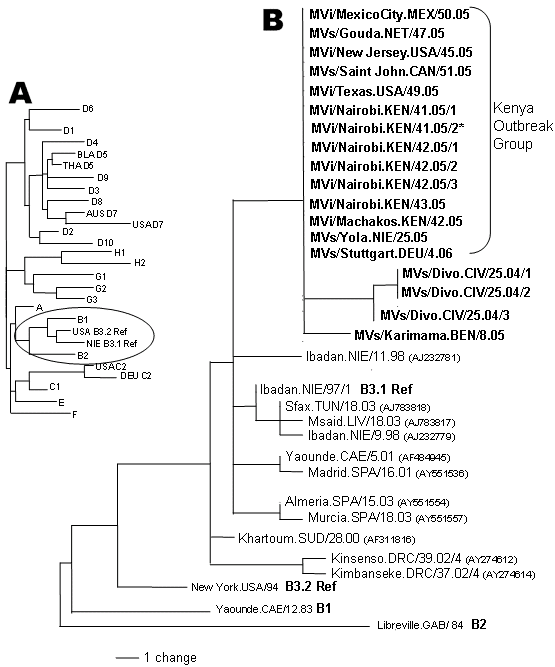
Figure. A) Dendrogram showing the relationships among the measles reference strains representing the 23 known measles genotypes (B3 has 2 reference strains). Clade B (circled) is expanded in panel B. B) Midpoint-rooted maximum parsimony tree of nucleoprotein genes (450 nt) of measles viruses from patients in the United States, Mexico, the Netherlands, Canada, and Kenya during 2005 and 2006. The unrooted tree includes sequences from Nigeria in 2005, Germany in 2006, Côte d'Ivoire in 2004, and Benin in 2005 (sequences in bold, this study) as well as selected B3 sequences available from GenBank for comparison. GenBank accession numbers are shown in parentheses. The identical sequences from the "Kenya Outbreak Group" are represented by GenBank accession number DQ888751, MVi/New Jersey.USA/45.05. The GenBank numbers for the sequence from Benin (BEN) and the 3 sequences from Côte d'Ivoire (CIV) are EF031461, EF031458, EF031459, and EF031460. *Collected from the Dagoretti area of Nairobi; the other Nairobi sequences were from cases in the Eastleigh area.
Main Article
Page created: October 14, 2011
Page updated: October 14, 2011
Page reviewed: October 14, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
