Volume 12, Number 11—November 2006
Dispatch
Molecular Characterization of Tickborne Relapsing Fever Borrelia, Israel
Figure 2
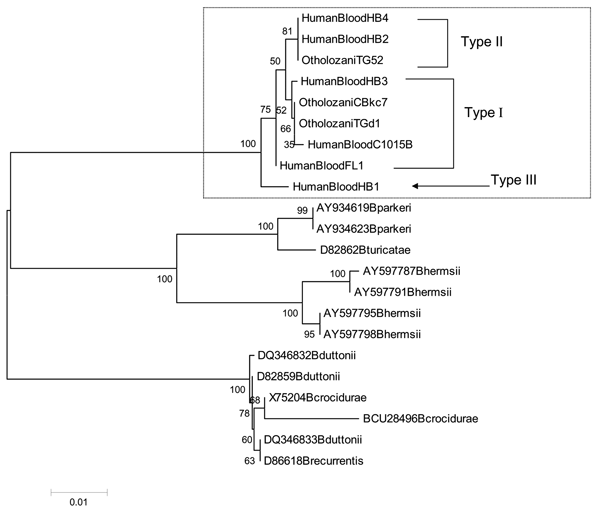
Figure 2. Phylogenetic tree based on flaB nucleotide sequences. The tree was constructed by the neighbor-joining method in a pairwise deletion procedure. Distances were calculated according to the Jukes and Cantor method. Numbers at nodes correspond to the percentage confidence level in a bootstrap test performed on 1,000 replicates. The scale bar corresponds to a 0.01 distance. The GenBank accession numbers for nucleotide sequences of Borrelia persica flaB shown here are as follows: HumanBloodFL1 (DQ673617), HumanBloodC1015B (DQ679904), OtholozaniCBkc7 (DQ679905), HumanBlood1 (DQ679906), HumanBlood2 (DQ679907), HumanBlood3 (DQ679908), HumanBlood4 (DQ679909), OtholozaniTG52 (DQ679910), and OtholozaniTGd1 (DQ679911).
Page created: October 17, 2011
Page updated: October 17, 2011
Page reviewed: October 17, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.