Volume 13, Number 11—November 2007
Dispatch
Novel Human Herpesvirus 8 Subtype D Strains in Vanuatu, Melanesia
Cite This Article
Citation for Media
Abstract
We show human herpesvirus 8 with diverse molecular subtype D variants to be highly endemic among the Ni-Vanuatu population. Most K1 genes were nearly identical to Polynesian strains, although a few clustered with Australian or Taiwanese strains. These results suggest diverse origins of the Ni-Vanuatu population and raise questions about the ancient human population movements in Melanesia.
Human herpesvirus 8 (HHV-8), or Kaposi Sarcoma Associated Herpesvirus (KSHV), is the etiologic agent of Kaposi sarcoma (KS). HHV-8 is not a widespread ubiquitous virus; its presence is mainly restricted to areas where classic or endemic KS is highly prevalent, i.e., estimates of HHV-8 seroprevalence in the general adult population range from 5% to >50% (1).
Exploiting the highly genetic variability of the HHV-8 K1 gene, molecular epidemiology led to the identification of 5 major K1 subtypes (A–E), some of which appear to be strongly linked to the geographic origin of the samples. Thus, the few known subtype D strains have been reported only in inhabitants from the Western Pacific region (2).
For people of Oceanian ancestry (including Melanesian, Polynesian, and Micronesian), very little data are available on the clinical and molecular epidemiology of HHV-8 and its associated diseases (3–9). Thus, we studied HHV-8 in the Vanuatu, an archipelago in the Southwest Pacific region, formerly named New Hebrides, which contains >80 islands (6 provinces). Indigenous Melanesians, also called Ni-Vanuatu, constitute 98% of the current population of ≈210,000. A recent study suggested that HHV-8 was rare in the Ni-Vanuatu population (10). Our goal for this cross-sectional study was to evaluate the prevalence of HHV-8 in the Vanuatu archipelago by using stringent serologic criteria and to characterize its genetic diversity.
Our work was performed on a large collection of ≈4,500 plasma and peripheral blood buffy coat (PBBC) samples from different islands of the archipelago, obtained in the framework of our previous studies on human T-cell lymphotropic virus (HTLV-1) (11,12). The field survey, carried out from April 2003 through August 2005, has been extensively described (11).
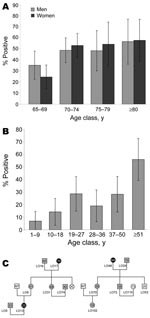
To detect plasma HHV-8 antibodies, an inhouse immunofluorescence assay (IFA) using BC-3 cells expressing only latent-associated nuclear antigens encoded by ORF73, was performed to detect plasma HHV-8 antibodies (13). Because HHV-8 seroprevalence increases with age in a virus-endemic population, we first tested a series of 376 samples, from persons >65 years of age (mean 72, median 70, range 65–96 years; 182 men and 194 women) originating from the 6 provinces of the archipelago (Appendix Figure). Among these 376 plasma samples, 170 (45.2%) were IFA positive at a 1:160 dilution, showing a clear typical nuclear spotted seroreactivity. The HHV-8 seroprevalence was similar between men (45.6%) and women (44.8%). The prevalence of HHV-8 increased with age, rising from 29.6% (65–69 years) to 57.1% (>80 years) (Figure 1, panel A) (p = 0.0005 trend χ2 test). This high level of HHV-8 seroprevalence was present in all 6 provinces (Appendix Figure).
A second serologic survey that used 237 plasma samples taken from 13 families with genealogic trees was performed (Figure 1, panels B and C). Among these 237 samples, 12 originated from elderly persons included in the 376 samples tested before. The HHV-8 seroprevalence was clearly age-dependent, rising from 6.9% among children 1–9 years of age to 28.2% in adults 50 years of age, followed by a new increase in persons >51 years of age (55.9%) (p<10–4 trend χ2 test). These results demonstrate for the first time, to our knowledge, that HHV-8 infection is endemic and circulates in the Ni-Vanuatu population.
We then characterized these HHV-8 strains molecularly. All DNA samples (1 μg), extracted from the PBBC, were first amplified by PCR for human β-globin sequences to control amplifiability. HHV-8 infection was determined by a nested PCR to obtain a 737-bp fragment of the open reading frame of the K1 gene (ORFK1). The first PCR was performed with the primer set K1AG75S/K1AG1200AS (14) and followed by a nested PCR with a second set of primers VR1S/VR2AS1 (15). All PCR products were purified from gel, cloned, and sequenced. Sequences were verified on both DNA strands. ORFK1 amplification was obtained from 32 (21.6%) of the 148 HHV-8–seropositive samples tested but in none of the 26 HHV-8–seronegative samples. Sequences were obtained for only 30 of the 32 ORFK1-positive PCRs (Table).
Comparative sequence analysis indicates that the 30 new sequences differed from each other. Furthermore, among them, 3 groups can be clearly identified. The first group comprises most strains (23/30) and corresponds to the sequences found in persons from the south central islands of the archipelago (Mallicolo, Ambrym, Epi, Tongoa, Emae, and Tanna); the second group comprises 4 sequences from persons living in the northern islands of Loh (LO13, LO17, LO48) and Santo (CESW32); and the third group involves only 3 sequences (ML10, ML36, ML46) from persons living in the northern island of Motalava.
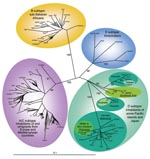
Phylogenetic analyses were performed on the 30 novel sequences obtained in this study, on all subtype D available K1 sequences, and on representatives of the different HHV-8 subtypes/subgroups, as described (4). The phylogenetic analyses were performed with all of the sequences available to date in GenBank. These sequences include 3 strains from Japan (J24, J25, and J26), 1 from Australia (3Au1), 1 from Taiwan (TKS10), 1 from New Zealand (ZKS3), and 1 from Wallis (WalKS1) (2,4, 6–8). Our results demonstrate that the Ni-Vanuatu HHV-8 clustered in 3 different genotype D subclades, which are highly supported phylogenetically with high bootstrap values of 99% or 100% (Figure 2). The first one comprising most strains corresponds to sequences closely related to each other and to the 2 Polynesian strains WalKS1 and ZKS3 (4,8). The second group (Loh/Santo), with only 4 sequences, was closely related to the Taiwanese strain TKS10 (8). The last 3 sequences from Motalava were nearly identical to the only strain from Australia (6).
Furthermore, phylogenetic analysis showed a star-like tree with a long branch for the Polynesian clade, which includes most Ni-Vanuatu strains. This finding strongly suggests a common origin or ancestor for these strains, with a possible founder effect (Figure 2). Based on stringent serologic and molecular analyses, our study demonstrates for the first time, to our knowledge, that HHV-8 infection is endemic in a Melanesian population.
Our serologic findings are consistent with those found in some remote villages of Papua New Guinea (5,9). On the basis of these studies, it is tempting to suggest that intrafamilial HHV-8 transmission occurs in such populations, as previously demonstrated in highly HHV-8 endemic populations of African origin (13).
From the molecular point of view, finding such a high molecular diversity of HHV-8 subtype D with some Polynesian-, Taiwanese- and Australian-like strains was surprising. These heterogeneous findings contrast with the more homogenous situation found for HTLV-1 genotypes in the same population (11).
Our molecular findings suggest that HHV-8 has been introduced in the Ni-Vanuatu populations by different migrations of infected persons. This conclusion is strengthened by the clustering of the Australian- and Taiwanese-like strains in the northern islands of Loh and Motalava. A variety of scenarios have been proposed to explain the peopling of near and remote Oceania, and our data highlight the possible multiple origins of Ni-Vanuatu ancestors. Ongoing molecular studies on both viral and mitochondrial/nuclear DNA will contribute to this debate through analyses of the variations observed. Indeed, these variations are intimately linked with the dispersal of early human settlers; analyses of the genetic variability of HHV-8 can help us reconstruct the patterns of human dispersal into Oceania (11).
Acknowledgments
We thank the Vanuatu Family Health Association, a local nongovernmental association, for its continuous support and interest in this work and Hélène Walter and Woreka Mera for their remarkable field work.
This study was financially supported by the Institut Pasteur, the Institut Pasteur de Nouvelle-Calédonie, and by the Centre National de la Recherche Scientifique. Renan Duprez was supported by a fellowship from the Caisse Nationale d’Assurance Maladie et Maternité des Travailleurs Non Salariés des Professions Non Agricoles.
Mr Cassar is a PhD candidate with primary research interest in the clinical and molecular epidemiology and physiopathology of dengue viruses. He is working on the epidemiology of HTLV-1 and HHV-8 in Melanesian populations.
References
- Dukers NH, Rezza G. Human herpesvirus 8 epidemiology: what we do and do not know. AIDS. 2003;17:1717–30. DOIPubMedGoogle Scholar
- Hayward GS, Zong JC. Modern evolutionary history of the human KSHV genome. Curr Top Microbiol Immunol. 2007;312:1–42. DOIPubMedGoogle Scholar
- Sengupta SK, Edwards K, Blair A, Hamilton DR, Niblet JS. Childhood Kaposi’s sarcoma in Papua New Guinea. Aust Paediatr J. 1986;22:301–4.PubMedGoogle Scholar
- Duprez R, Cassar O, Hbid O, Rougier Y, Morisse L, Bassot S, Cutaneous disseminated endemic Kaposi’s sarcoma in a Polynesian man infected with a new divergent human herpesvirus 8 subtype D. J Clin Virol. 2006;37:222–6. DOIPubMedGoogle Scholar
- Rezza G, Danaya RT, Wagner TM, Sarmati L, Owen IL, Monini P, Human herpesvirus-8 and other viral infections, Papua New Guinea. Emerg Infect Dis. 2001;7:893–5. DOIPubMedGoogle Scholar
- Meng YX, Spira TJ, Bhat GJ, Birch CJ, Druce JD, Edlin BR, Individuals from North America, Australasia, and Africa are infected with four different genotypes of human herpesvirus 8. Virology. 1999;261:106–19. DOIPubMedGoogle Scholar
- Meng YX, Sata T, Stamey FR, Voevodin A, Katano H, Koizumi H, Molecular characterization of strains of human herpesvirus 8 from Japan, Argentina and Kuwait. J Gen Virol. 2001;82:499–506.PubMedGoogle Scholar
- Zong JC, Ciufo DM, Alcendor DJ, Wan X, Nicholas J, Browning PJ, High-level variability in the ORF-K1 membrane protein gene at the left end of the Kaposi's sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations. J Virol. 1999;73:4156–70.PubMedGoogle Scholar
- Suligoi B, Danaya RT, Sarmati L, Owen IL, Boros S, Pozio E, Infection with human immunodeficiency virus, herpes simplex virus type 2, and human herpesvirus 8 in remote villages of southwestern Papua New Guinea. Am J Trop Med Hyg. 2005;72:33–6.PubMedGoogle Scholar
- Satoh M, Kaneko A, Kokaze A, Katano H, Sata T. Seroprevalence of human herpesvirus 8 in the Vanuatu islands in eastern Melanesia. Jpn J Infect Dis. 2006;59:63–5.PubMedGoogle Scholar
- Cassar O, Capuano C, Bassot S, Charavay F, Duprez R, Afonso PV, HTLV-1 Subtype C Melanesian genetic variants of Vanuatu Archipelago and Solomon Islands share a common ancestor. J Infect Dis. 2007;196:510–21. DOIPubMedGoogle Scholar
- Cassar O, Capuano C, Meertens L, Chungue E, Gessain A. Human T-cell leukemia virus type 1 molecular variants, Vanuatu, Melanesia. Emerg Infect Dis. 2005;11:706–10.PubMedGoogle Scholar
- Plancoulaine S, Abel L, van Beveren M, Gessain A. High titers of anti-human herpesvirus 8 antibodies in elderly males in an endemic population. J Natl Cancer Inst. 2002;94:1333–5.PubMedGoogle Scholar
- Lacoste V, Judde JG, Brière J, Tulliez M, Garin B, Kassa-Kelembho E, Molecular epidemiology of human herpesvirus 8 in africa: both B and A5 K1 genotypes, as well as the M and P genotypes of K14.1/K15 loci, are frequent and widespread. Virology. 2000;278:60–74. DOIPubMedGoogle Scholar
- Kadyrova E, Lacoste V, Duprez R, Pozharissky K, Molochkov V, Huerre M, Molecular epidemiology of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 strains from Russian patients with classic, posttransplant, and AIDS-associated Kaposi's sarcoma. J Med Virol. 2003;71:548–56. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleTable of Contents – Volume 13, Number 11—November 2007
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Antoine Gessain, Unité d’Epidémiologie et Physiopathologie des Virus Oncogènes/URA CNRS 3015, Département de Virologie, Bâtiment Lwoff, Institut Pasteur, 28 rue du Dr. Roux, 75724 Paris CEDEX 15, France;
Top