Volume 15, Number 3—March 2009
Letter
Cat-to-Human Orthopoxvirus Transmission, Northeastern Italy
Figure
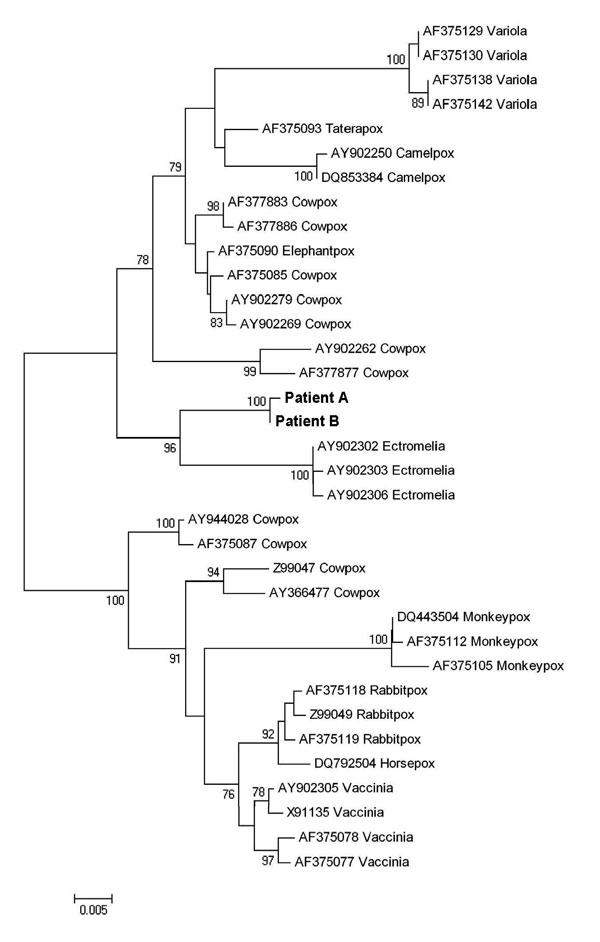
Figure. Phylogenetic tree of nucleotide sequences of the complete hemagglutinin open reading frame (930 bp) of orthopoxviruses (OPVs) isolated from the 2 patients described in this report and additional poxviruses available in GenBank (cowpox: AY902279, AY902269, AF375085, AF377883, AF377886, AY902262, AY944028, Z99047, AY366477, AF377877, AF375087; taterapox: AF375093; camelpox: AY902250, DQ853384; horsepox: DQ792504; elephantpox: AF375090; vaccinia: AY902305, X91135, AF375078, AF375077; rabbitpox: AF375119, AF375118, Z99049; variola: AF375129, AF375130, AF375138, AF375142; ectromelia: AY902302, AY902303, AY902306; monkeypox: DQ443504, AF375105, AF375112). Multiple alignment was generated with ClustalW 1.7 software in BioEdit (www.mbio.ncsu.edu/BioEdit/BioEdit.html), and the phylogenetic tree was constructed by using maximum-likelihood and neighbor-joining algorithms implemented in Mega 4.0 software (www.megasoftware.net). Bootstrap values >75 are shown at nodes. The sequences from the patients described in this report form a distinct cluster separate from other OPV sequences. Scale bar indicates genetic distance.