Volume 15, Number 8—August 2009
Letter
KI and WU Polyomaviruses in Patients Infected with HIV-1, Italy
Figure
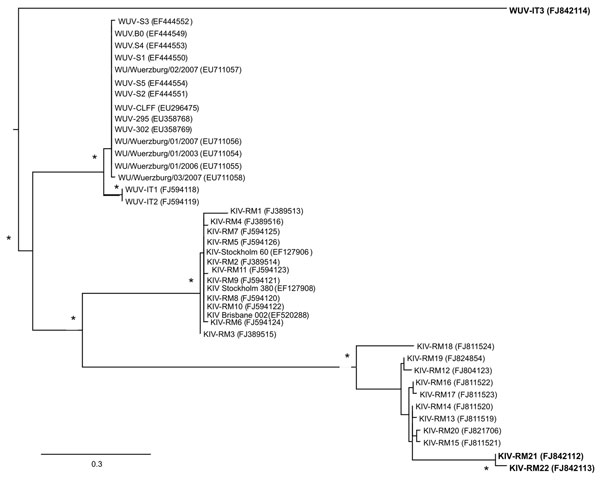
Figure. Unrooted phylogenetic tree showing analysis of KI (KIV-RM21, KIV-RM22) and WU (WUV-IT3) polyomaviruses (KIPyVs, WUPyVs, respectively) identified in the plasma of HIV-1–positive patients. The identified strains are indicated in boldface, and the phylogenetic analysis refers to the small t region. The other polyomaviruses shown in the figure are the KIPyVs (KIV-RM1 to KIV-RM20) and WUPyVs (WUV-IT1 and WUV-IT2) identified in Italy in previous studies (6,7) and the prototype strains for KIPyV (GenBank accession nos. EF127906, EF127908, EF520288) and WUPyV (GenBank accession nos. EF444549–EF444554, EU711054–EU711058, EU296475, EU358768, and EU358769). GenBank accession numbers for all virus strains are shown in parentheses. Multiple nucleotide sequence alignments were performed by using ClustalX software (http://bips.u-strasbg.fr/fr/documentation/clustalx/#g), and the phylogenetic tree was constructed by using the neighbor-joining algorithm with LogDet-corrected distances (http://paup.csit.fsu.edu/about.html) (8). An asterisk (*) beside a branch represents significant statistical support for the clade subtending that branch (p<0.001 in the zero-branch–length test) and bootstrap support >75%. Scale bar indicates nucleotide substitutions per site.
References
- Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet. 1971;1:1257–60.DOIGoogle Scholar
- Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (BK) isolated from urine after renal transplantation. Lancet. 1971;1:1253–7.DOIGoogle Scholar
- Allander T, Andreasson K, Gupta S, Bjerkner M, Bogdanovic G, Persson MA, Identification of a third human polyomavirus. J Virol. 2007;81:4130–7.DOIGoogle Scholar
- Gaynor AM, Nissen MD, Whiley DM, McKay IM, Lambert SB, Wu G, Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 2007;3:e64.DOIGoogle Scholar
- Norja P, Ubillos I, Templeton K, Simmonds P. No evidence for an association between infections with WU and KI polyomaviruses and respiratory disease. J Clin Virol. 2007;40:307–11.DOIGoogle Scholar
- Babakir-Mina M, Ciccozzi M, Campitelli L, Aquaro S, Lo Coco A, Perno CF, Identification of the novel KI polyomavirus in paranasal and lung tissues. J Med Virol. 2009;81:558–61.DOIGoogle Scholar
- Babakir-Mina M, Ciccozzi M, Alteri C, Polchi P, Picardi A, Greco F, Excretion of the novel polyomaviruses KI and WU in the stool of patients with hematological disorders. J Med Virol. 2009. In press.
- Neske F, Blessing K, Pröttel A, Ullrich F, Kreth HW, Weissbrich B. Detection of WU polyomavirus DNA by real-time PCR in nasopharyngeal aspirates, serum, and stool samples. J Clin Virol. 2009;44:115–8.DOIGoogle Scholar
- Venter M, Visser A, Lassauniere R. Human polyomaviruses, WU and KI in HIV-exposed children with acute lower respiratory tract infections in hospitals in South Africa. J Clin Virol. 2009;44:230–4.DOIGoogle Scholar
- Mourez T, Bergeron A, Ribaud P, Scieux C, de Latour RP, Tazi A, Polyomaviruses KI and WU in immunocompromised patients with respiratory disease. Emerg Infect Dis. 2009;15:107–9.DOIGoogle Scholar