Volume 16, Number 1—January 2010
Dispatch
Broiler Chickens as Source of Human Fluoroquinolone-Resistant Escherichia coli, Iceland
Abstract
To investigate feed as a source for fluoroquinolone-resistant Escherichia coli in broiler chickens, we compared antimicrobial drug–resistant E. coli from broiler feed and broilers with ciprofloxacin-resistant human clinical isolates by using pulsed-field gel electrophoresis. Feed was implicated as a source for ciprofloxacin-resistant broiler-derived E. coli and broilers as a source for ciprofloxacin-resistant human-derived E. coli.
In a previous study, we found a relatively high prevalence of antimicrobial and especially quinolone resistance among Escherichia coli isolates from broiler chickens and broiler meat (1), despite no known antimicrobial drug selection pressure in chicken farming in Iceland and biosecurity measures to prevent transmission of infectious agents into farms. Broiler houses are cleaned and disinfected after broiler flocks are transported to slaughter. Therefore, resistant bacteria are unlikely to persist in the broiler houses. However, animal feed can be contaminated with antimicrobial drug–resistant E. coli (2).
The high prevalence of quinolone-resistant E. coli isolates obtained from broilers and broiler meat coincides with an increasing prevalence of fluoroquinolone resistance among human clinical E. coli isolates in Iceland. This increase correlated with increased use of fluoroquinolones in clinical settings (3).
We examined whether the prevalence of resistant E. coli strains in broilers had changed since our previous study and whether broiler feed could be a source for the resistant strains. Furthermore, we compared the genotypes of ciprofloxacin-resistant broiler, broiler meat, and broiler feed E. coli isolates with ciprofloxacin-resistant human clinical E. coli isolates.
The sampling period for this study was May through November 2008. Pooled cecal samples (20 ceca from each flock) were taken from the 30 flocks slaughtered at all 3 broiler slaughterhouses in Iceland in June 2008. Ceca were stomachered in phosphate-buffered saline, spread on MacConkey agar with and without enrofloxacin (0.25 mg/L), and incubated overnight. Feed was sampled from feed stalls at 18 farms (of which 14 had participated in the previous study) and from 2 feed mills; the feed was suspended in buffered peptone water, mixed, incubated overnight, and spread on MacConkey agar as described above. One colony from each agar plate was selected for susceptibility testing as described in our previous study (1). Because isolates were collected from the 18 largest broiler farms (of the 27 farms operating in Iceland), all the broiler slaughterhouses, and the only 2 feed mills operating in Iceland, we believe this study provides a representative sample.
We selected all 34 available human E. coli isolates recovered from routine clinical specimens (mostly urine and blood) at the main clinical microbiology/reference laboratory in Iceland (Landspitali University Hospital) during 2006–2007, which had similar susceptibility patterns to the strains previously isolated from broilers (1) (ampicillin-tetracycline-sulfamethoxazole/trimethoprim-ciprofloxacin or ciprofloxacin alone). Only 1 isolate was chosen from each patient.
We performed susceptibility testing using a microbroth dilution method (VetMIC; National Veterinary Institute, Uppsala, Sweden). MICs were determined for ampicillin, cefotaxime, ceftiofur, chloramphenicol, ciprofloxacin, florfenicol, gentamicin, nalidixic acid, kanamycin, streptomycin, sulfamethoxazole, tetracycline, and trimethoprim. Cutoff values were those used in the monitoring programs in Sweden and Norway (4,5). Strains resistant to >3 classes of antimicrobial agents were considered multiresistant.
We compared all E. coli broiler ceca and feed isolates resistant to >1 antimicrobial agents and the 34 ciprofloxacin-resistant human E. coli isolates with resistant E. coli isolates from the previous study (2005–2007) (1) by pulsed-field gel electrophoresis (PFGE) using a slightly modified method of Ribot et al. (6). Comparison of PFGE patterns was made by visual inspection and BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium). For cluster analysis, the Dice coefficient for band matching (band-position tolerance 1.5%) was used to generate an unweighted pair group method with arithmetic averages dendrogram. Isolates from the previous and present study that did not yield a satisfactory banding pattern by PFGE were genotyped by randomly amplified polymorphic DNA (RAPD) analysis as described (7). Reaction products were analyzed by electrophoresis on 1.5% agarose gels stained with ethidium bromide. Patterns were considered different when the profiles differed by at least 1 band. Similarity among RAPD patterns was compared as described for PFGE. Clusters for PFGE and composite RAPD profiles were defined as >2 isolates with >80% similarity. Prevalence values were compared by using the Fisher exact test.
Of the 40 broiler isolates, 20 were resistant to >1 of the antimicrobial drugs tested (Table); only 1 was multidrug resistant (resistant to streptomycin, tetracyclin, sulfamethoxazole, and trimethoprim). Ciprofloxacin and nalidixic acid were always cross-resistant. Compared with the previous sampling, the prevalence of resistance increased significantly for ciprofloxacin and nalidixic acid (from 18.2% to 42.5%; p<0.0001) but decreased significantly for ampicillin (from 18.2% to 0.0%; p = 0.002) and sulfamethoxazole (from 19.1% to 5.0%; p = 0.0398). This finding suggests that quinolone resistance was not transferred with resistance to the other antimicrobial agents and that it was selected for by other factors. Of the 22 E. coli isolates obtained from feed, 7 (32%) were resistant to ciprofloxacin and nalidixic acid, and all were susceptible to the other antimicrobial agents tested. Although no E. coli were isolated from the 2 feed mill samples, other Enterobacteriaceae grew on the agar plates, which could have overgrown existing E. coli strains, if any, demonstrating that the feed was not sterile.
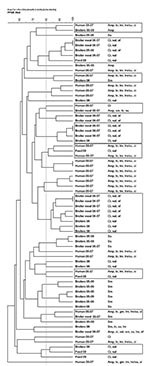
The 27 resistant broiler and feed isolates were compared with 76 resistant isolates analyzed in our previous study (1) along with the 34 ciprofloxacin-resistant human E. coli isolates. Of 137 broiler, broiler meat, feed and human isolates, 110 (80%) yielded interpretable, reproducible PFGE patterns. We detected 92 profiles, of which 81 (88%) were represented by a single isolate. Isolates of different origin were intermixed forming 26 clusters, of which 12 were seen in the previous study. Of the 14 new clusters, 10 contained isolates of different origins (Figure). Human isolates clustered with broiler (2005–2006), broiler meat, broiler (2008), and feed isolates in 6 clusters (Figure). This supports previous findings of chickens and their products as a possible source of antimicrobial drug–resistant E. coli in humans (8,9). With the extensive genomic diversity of E. coli and the discriminative power of PFGE typing, finding indistinguishable isolates of different origin collected over several years is unlikely, except from a large collection (8,10). Therefore, finding human isolates closely related (>80 similarity) to broiler, broiler meat, and feed isolates suggests an epidemiologic link between the populations. Additionally, we found closely related isolates from feed and broiler (samples from 2008 and 2005–2006) from geographically distant farms, supporting previous findings that antimicrobial drug–resistant E. coli could be introduced into the farm environment through broiler feed (2).
The isolates that did not give interpretable PFGE patterns (1 broiler [2005–2006] and 4 broiler meat [2006–2007] isolates, 11 broiler [2008] and feed [2008] isolates, and 11 ciprofloxacin-resistant human isolates) were subjected to RAPD analysis. All isolates yielded interpretable patterns displaying 26 distinct profiles; all but 1 unique profile represented a single isolate. At 80% similarity, 1 cluster was of mixed origin, containing 6 isolates from feed and broilers (2008).
Prevalence of fluoroquinolone-resistant E. coli remains moderately high in broilers, but resistance to other antimicrobial drugs is decreasing. Fluoroquinolone-resistant E. coli isolated from broiler feed implicates feed as the source of resistant strains into farms. Resistant isolates from feed, broilers, broiler meat, and humans were closely related, demonstrating that poultry and their food products can be a source of resistant E. coli in humans.
Ms Thorsteinsdottir is a biologist and a PhD fellow at the Institute for Experimental Pathology, University of Iceland, Keldur. Her primary research interest is antimicrobial drug resistance of bacteria, particularly in relation to zoonoses.
Acknowledgment
The study was funded by the University of Iceland Research Fund and The Icelandic Research Fund for Graduate Students.
References
- Thorsteinsdottir TR, Haraldsson G, Fridriksdottir V, Kristinsson KG, Gunnarsson E. Prevalence and genetic relatedness of antimicrobial resistant Escherichia coli isolated from animals, foods and humans in Iceland. Zoonoses Public Health. 2009 Nov 13; [Epub ahead of print].
- da Costa PM, Oliveira M, Bica A, Vaz-Pires P, Bernardo F. Antimicrobial resistance in Enterococcus spp. and Escherichia coli isolated from poultry feed and feed ingredients. Vet Microbiol. 2007;120:122–31. DOIPubMedGoogle Scholar
- Jonsdottir K, Kristinsson KG. Quinolone resistance in gram-negative rods in Iceland and association with antibiotic use [in Icelandic]. Laeknabladid. 2008;94:279–85.PubMedGoogle Scholar
- NORM/NORM-VET 2004. Usage of antimicrobial agents and occurrence of antimicrobial resistance in Norway. Tromsø/Oslo (Norway): National Veterinary Institute; 2004.
- SVARM. 2006. Swedish veterinary antimicrobial resistance monitoring. Uppsala (Sweden): National Veterinary Institute; 2007.
- Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3:59–67. DOIPubMedGoogle Scholar
- Pacheco AB, Guth BE, Soares KC, Nishimura L, de Almeida DF, Ferreira LC. Random amplification of polymorphic DNA reveals serotype-specific clonal clusters among enterotoxigenic Escherichia coli strains isolated from humans. J Clin Microbiol. 1997;35:1521–5.PubMedGoogle Scholar
- Johnson JR, Kuskowski MA, Menard M, Gajewski A, Xercavins M, Garau J. Similarity between human and chicken Escherichia coli isolates in relation to ciprofloxacin resistance status. J Infect Dis. 2006;194:71–8. DOIPubMedGoogle Scholar
- Al-Ghamdi MS, El-Morsy F, Al-Mustafa ZH, Al-Ramadhan M, Hanif M. Antibiotic resistance of Escherichia coli isolated from poultry workers, patients and chicken in the eastern province of Saudi Arabia. Trop Med Int Health. 1999;4:278–83. DOIPubMedGoogle Scholar
- Ramchandani M, Manges AR. DebRoy C, Smith SP, Johnson JR, Riley LW. Possible animal origin of human-associated, multidrug-resistant, uropathogenic Escherichia coli. Clin Infect Dis. 2005;40:251–7. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleTable of Contents – Volume 16, Number 1—January 2010
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Thorunn R. Thorsteinsdottir, Institute for Experimental Pathology, University of Iceland, Keldur v/Vesturlandsveg, 112 Reykjavík, Iceland
Top