Volume 16, Number 12—December 2010
Dispatch
Leishmania tropica Infection in Golden Jackals and Red Foxes, Israel
Figure
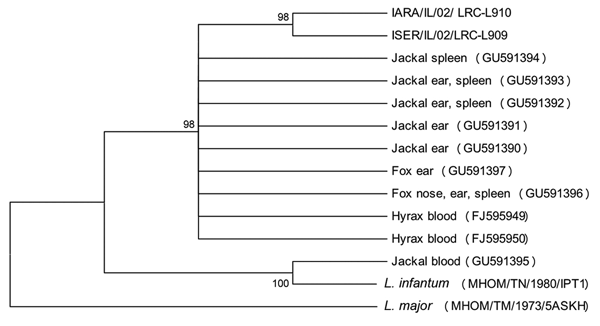
Figure. Neighbor-joining tree phylogram comparing internal transcribed spacer 1 (ITS1) Leishmania tropica DNA sequences from wild canids, Israel. The neighbor-joining tree constructed in MEGA version 3.0 (www.megasoftware.net) by the ITS1 HRM PCR sequences (222–239 nt) agrees with the maximum-likelihood algorithm. The tree shown is based on the Kimura 2-parameter model of nucleotide substitution. Bootstrap values are based on 1,000 replicates. The analysis provided tree topology only; the lengths of the vertical and horizontal lines are not significant. L. major was used as an outgroup. GenBank accession numbers of L. tropica from hyraxes deposited from this study are shown in brackets. Numbers on nodes represent bootstrap values. MHOM, human; IARA, Phlebotomus arabicus sand fly; ISER, Phlebotomus sergenti sandfly.