Volume 16, Number 9—September 2010
Dispatch
KI and WU Polyomaviruses and CD4+ Cell Counts in HIV-1–infected Patients, Italy
Cite This Article
Citation for Media
Abstract
To investigate an association between KI and WU polyomavirus (KIPyV and WUPyV) infections and CD4+ cell counts, we tested HIV-1–positive patients and blood donors. No association was found between cell counts and virus infections in HIV-1–positive patients. Frequency of KIPyV infection was similar for both groups. WUPyV was more frequent in HIV-1–positive patients.
BK and JC polyomaviruses are known to infect humans (1,2). Recently, the novel KI polyomavirus (KIPyV) and WU polyomavirus (WUPyV) have been identified in respiratory secretions of children with signs of acute respiratory disease (3,4). However, there is little evidence that these viruses are the causative agents of respiratory disease. The pathogenic role of these viruses in immunocompromised patients is also unclear.
In a study that investigated human polyomaviruses in autopsy lymphoid tissue samples from patients who were positive for HIV, KIPyV was detected in 7.1% of immunocompromised patients with AIDS and in 1.8% of nonimmunocompromised controls; WUPyV was detected in 9.5% of patients with AIDS but not in controls (5). We detected KIPyV and WUPyV in 3.2% and 1.6%, respectively, of plasma samples from HIV-1–infected patients (6). To determine an association between infection with KIPyV and WUPyV and CD4+ cell counts, we obtained plasma samples from HIV-1–positive patients having high and low CD4+ cell counts and a group of healthy controls and tested them for these 2 polyomaviruses.
Plasma specimens from 153 HIV-1–infected persons (75% male patients, median age 41.9 years, interquartile range 33.8–47.3 years) with high (110 persons) and low (43 persons) CD4+ counts and from 130 blood donors (80% male donors, median age 41 years, interquartile range 32–47.5 years) were obtained at the Foundation Polyclinic Tor Vergata in Rome, Italy, during 2004–2009. Of 153 HIV-1–infected patients, 74 were receiving highly active antiretroviral therapy: a nucleoside reverse transcriptase inhibitor (NRTI) and a protease inhibitor (PI) (n = 35 patients); an integrase inhibitor (INI), an NRTI, and a PI (n = 7); a nonnucleoside-reverse transcriptase inhibitor and an NRTI (n = 26); an INI and an NRTI (n = 2); an INI and an NNRTI (n = 2); a chemokine receptor type 5 antagonist, an NRTI, and a PI (n = 1); and a chemokine receptor type 5 antagonist, an INI, and a nonnucleoside-reverse transcriptase inhibitor (n = 1). Sixty patients did not receive any therapy. No information was available for 19 patients. Additional information available for patients included HIV-1 viremia and co-infection with hepatitis B virus and hepatitis C virus.
Phylogenetic analysis of the small T antigen gene of KIPyV and WUPyV was performed as described (6,7). GenBank accession numbers of the sequences used in this analysis are shown in the Table A1.
Total DNA was extracted from 0.2-mL plasma samples by using QIAamp DNA Mini Kit (QIAGEN, Milan, Italy) according to the manufacturer’s instructions and stored at –80°C until analysis. Amplification of KIPyV and WUPyV was conducted as described (8,9). A standard curve was created in a 4-log range by using 1:10 serial dilutions of a virus-specific standard. The dynamic range was determined by using 10-fold dilutions (1010–100 copies/reaction) of each sample. Sensitivity of the 2 methods, which corresponded to the lowest plasma dilution detectable at a frequency of 100%, was evaluated. The dynamic range was 102–1010 for KIPyV and 101–1010 for WUPyV.
Statistical analysis was performed by using Epi Info version 3.5.1 software (Centers for Disease Control and Prevention, Atlanta, GA, USA). Odds ratios were determined for associations between infection with HIV and infection with KIPyV and WUPyV and other variables. Statistical significance was assessed by calculating 95% confidence intervals (CIs) and by using standard nonparametric statistics.
Real-time PCR detected KIPyV and WUPyV in 4 (2.6%) of 153 and 7 (4.6%) of 153 HIV-1–infected patients, respectively (Table 1). Of the 130 blood donors examined, 4 and 1 were positive for KIPyV (3.1%) and WUPyV (0.8%), respectively. For KIPyV, no difference was detected in the frequency of infection between HIV-1–infected patients and blood donors. Patients infected with HIV-1 had a higher risk for infection with WUPyV infection than did blood donors. However, this difference showed borderline statistical significance (odds ratio 6.15, 95% CI 0.93–141; p = 0.054). For WUPyV-positive and KIPyV-positive patients, median CD4+ cell counts were 308 cells/µL (95% CI 248–523 cells/µL) and 356 cells/µL (95% CI 270–517 cells/µL), respectively. No association was observed between CD4+ cell counts and risk for infection with KIPyV or WUPyV.
Median HIV-1 virus load in persons infected with WUPyV or KIPyV was 3,210 copies/mL (95% CI 108–36,895 copies/mL) or 60,636 copies/mL (95% CI 2,791–246,500 copies/mL), respectively. When HIV-1 virus load, CD4+ cell count, and co-infection with hepatitis B and C viruses were analyzed in patients infected with KIPyV or WUPyV, no association was found. Of 11 patients infected with KIPyV or WUPyV, 6 received highly active antiretroviral therapy and 5 did not receive any therapy (Table 2).
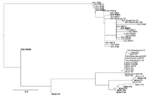
Phylogenetic analysis showed that all WUPyVs identified in this study except WUV-IT4 are closely related to the WUV-IT3 strain identified in an HIV-1 patient (6) (Figure). The KIPyV strains identified are relatively distant from those identified in another study (6), except for strain KIV-RM23, which clusters with KIV-RM21 (6) (Figure).
KIPyV and WUPyV have been identified in respiratory secretions of pediatric patients (3,4). New polyomaviruses have also been detected in immunocompromised patients (10–13). However, the pathogenic role of these polyomaviruses in immunocompromised patients is unclear. No associations were found between CD4+ cell counts in HIV-1–positive patients and infection with KIPyV or WUPyV. Frequency of KIPyV infection for HIV-1–positive patients was similar to that for blood donors. However, frequency of WUPyV infection was higher for HIV-1–positive patients than for blood donors, although this difference showed borderline significance.
Detection of WUPyV did not show a correlation with virus load for HIV-1 or lower CD4+ cell counts. Seroprevalence of KIPyV and WUPyV in an adult population was 55% and 69%, respectively (14). The higher rate of infection for WUPyV may account for the higher rate of detection for WUPyV in our study population. In a previous study (6), prevalence of WUPyV in plasma of HIV-1–positive patients was lower than that in our study. This difference may have been caused by the larger sample size in our study.
Phylogenetic analysis did not suggest circulation of specific KIPyV and WUPyV strains in HIV-1–positive patients. The KIPyVs identified in this study cluster with those identified in HIV-1–positive patients, and the WUPyVs identified are closely related to the strain identified previously in an HIV-1–positive patient (6). However, these WUPyV strains also cluster with strain WUV-IT1 and 2 strains identified in stool samples (10).
Our study design and the complex nature of AIDS-related disease do not enable one to make definitive conclusions on the role of novel polyomaviruses in HIV-1–positive patients. However, our data seem to exclude an active role for KIPyV and WUPyV in HIV-1–positive patients.
We detected WUPyV and KIPyV in healthy persons and immunocompromised persons. BK and JC polyomaviruses persist in peripheral blood mononuclear cells in healthy persons (15). However, frequency of detection may vary from 0% to 90% of persons tested. This large variation may reflect recent infection or virus reactivation in a subgroup of persons (15). Thus, detection of KIPyV and WUPy in blood cells of immunocompetent persons is needed to identify a possible hematologic reservoir.
Dr Babakir-Mina is a research scientist at the Foundation University Hospital Tor Vergata in Rome, Italy. His research interests are molecular diagnosis of polyomaviruses and their role in immunocompromised and healthy persons.
Acknowledgment
We thank Romina Salpini, Paola Galati, and Francesca Stazi for obtaining HIV-positive samples.
References
- Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet. 1971;1:1257–60. DOIPubMedGoogle Scholar
- Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (BK) isolated from urine after renal transplantation. Lancet. 1971;1:1253–7. DOIPubMedGoogle Scholar
- Allander T, Andreasson K, Gupta S, Bjerkner M, Bogdanovic G, Persson MA, Identification of a third human polyomavirus. J Virol. 2007;81:4130–7. DOIPubMedGoogle Scholar
- Gaynor AM, Nissen MD, Whiley DM, McKay IM, Lambert SB, Wu G, Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 2007;3:e64. DOIPubMedGoogle Scholar
- Sharp CP, Norja P, Anthony I, Bell JE, Simmonds P. Reactivation and mutation of newly discovered WU, KI, and Merkel cell carcinoma polyomaviruses in immunosuppressed individuals. J Infect Dis. 2009;199:398–404. DOIPubMedGoogle Scholar
- Babakir-Mina M, Ciccozzi M, Trento E, Perno CF, Ciotti M. KI and WU polyomaviruses in patients infected with HIV-1, Italy. Emerg Infect Dis. 2009;15:1323–5. DOIPubMedGoogle Scholar
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–8. DOIPubMedGoogle Scholar
- Babakir-Mina M, Ciccozzi M, Bonifacio D, Bergallo M, Costa C, Cavallo R, Identification of the novel KI and WU polyomaviruses in human tonsils. J Clin Virol. 2009;46:75–9. DOIPubMedGoogle Scholar
- Bergallo M, Terlizzi ME, Astegiano S, Ciotti M, Babakir-Mina M, Perno CF, Real time PCR TaqMan assays for detection of polyomaviruses KIV and WUV in clinical samples. J Virol Methods. 2009;162:69–74. DOIPubMedGoogle Scholar
- Babakir-Mina M, Ciccozzi M, Alteri C, Polchi P, Picardi A, Greco F, Excretion of the novel polyomaviruses KI and WU in the stool of patients with hematological disorders. J Med Virol. 2009;81:1668–73. DOIPubMedGoogle Scholar
- Venter M, Visser A, Lassauniere R. Human polyomaviruses, WU and KI in HIV exposed children with acute lower respiratory tract infections in hospitals in South Africa. J Clin Virol. 2009;44:230–4. DOIPubMedGoogle Scholar
- Mourez T, Bergeron A, Ribaud P, Scieux C, de Latour RP, Tazi A, Polyomaviruses KI and WU in immunocompromised patients with respiratory disease. Emerg Infect Dis. 2009;15:107–9. DOIPubMedGoogle Scholar
- Barzon L, Squarzon L, Militello V, Trevisan M, Porzionato A, Macchi V, WU and KI polyomaviruses in the brains of HIV-positive patients with and without progressive multifocal leukoencephalopathy. J Infect Dis. 2009;200:1755–8. DOIPubMedGoogle Scholar
- Kean JM, Rao S, Wang M, Garcea RL. Seroepidemiology of human polyomaviruses. PLoS Pathog. 2009;5:e1000363. DOIPubMedGoogle Scholar
- Dolei A, Pietropaolo V, Gomes E, Di Taranto C, Ziccheddu M, Spanu MA, Polyomavirus persistence in lymphocytes: prevalence in lymphocytes from blood donors and healthy personnel of a blood transfusion centre. J Gen Virol. 2000;81:1967–73.PubMedGoogle Scholar
Figure
Tables
Cite This ArticleTable of Contents – Volume 16, Number 9—September 2010
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Marco Ciotti, Foundation University Hospital Tor Vergata, Viale Oxford 81, 00133 Rome, Italy
Top