Volume 16, Number 9—September 2010
Research
Legionellosis Outbreak Associated with Asphalt Paving Machine, Spain, 2009
Cite This Article
Citation for Media
Abstract
From 1999 through 2005 in Alcoi, Spain, incidence of legionellosis was continually high. Over the next 4 years, incidence was lower, but an increase in July 2009 led health authorities to declare an epidemic outbreak. A molecular epidemiology investigation showed that the allelic profiles for all Legionella pneumophila samples from the 2009 outbreak patients were the same, thus pointing to a common genetic origin for their infections, and that they were identical to that of the organism that had caused the previous outbreaks. Spatial-temporal and sequence-based typing analyses indicated a milling machine used in street asphalt repaving and its water tank as the most likely sources. As opposed to other machines used for street cleaning, the responsible milling machine used water from a natural spring. When the operation of this machine was prohibited and cleaning measures were adopted, infections ceased.
Legionella pneumophila (1) is a gram-negative bacterium identified as the causative agent of an outbreak of pneumonia that occurred in a Philadelphia hotel during a Legionnaires’ convention in 1977 (2). This outbreak affected 221 persons, of whom 34 died (2). Although other Legionella spp. can cause the disease, L. pneumophila is responsible for 90% of the cases of legionellosis globally. This species is 1 of the most common causes of community-acquired bacterial pneumonia (3–7) and the second most common cause of severe pneumonia (8).
L. pneumophila is a waterborne bacterium that can cause respiratory illness when a susceptible person inhales contaminated, aerosolized water. Infection sources are usually human-made aquatic habitats, such as potable water supplies (9), whirlpool spas (10), cooling towers (11,12), showers (13), decorative fountains (14,15), and hoses (16).
Molecular epidemiologic analyses of L. pneumophila usually compare sequence-based typing patterns from bacterial cultures derived from putative environmental sources with L. pneumophila cultures from patients’ sputum samples. This approach consists of amplifying and sequencing internal fragments of 7 loci and assigns a number to the different alleles derived from each locus (17,18). The combination of allele variants for each locus determines the allelic profile that characterizes each sample. Isolation of L. pneumophila from respiratory secretions on selective media is fastidious and time-consuming. Moreover, some L. pneumophila strains may be viable but cannot be cultured (19); despite high specificity of this method, culturing L. pneumophila is not efficient (20,21). Therefore, the efficiency of sequence-based typing of L. pneumophila can be improved by direct amplification and sequencing of DNA extracted from uncultured respiratory samples (22).
Since 2000, a specific epidemiologic surveillance system for legionellosis has been in place in the Hospital Virgen de los Llirios in Alcoi, Spain. Every patient with signs of pneumonia is scanned by chest radiography and urine analysis for L. pneumophila serogroup 1 antigen. This surveillance system enables early detection and better prognosis for L. pneumophila–infected patients. It also helps distinguish between sporadic cases and outbreaks, thus enabling early start of epidemiologic investigations.
At the end of July 2009, the epidemiology surveillance system detected 2 cases of legionellosis in persons who had stayed in Alcoi, Spain, during their incubation periods. New cases appeared during the first week of August, at which time an epidemic outbreak was declared and an epidemiologic investigation was started. Patients in the outbreak were questioned about clinical and personal aspects. Spatial–temporal analysis was used to identify the most likely areas of exposure for infection. L. pneumophila was isolated from environmental samples obtained in those areas. Because the usual facilities and municipal water systems associated with risk were not contaminated, other facilities not previously linked to legionellosis outbreaks were considered. To verify the common genetic origin of the outbreak and its environmental source, we performed an epidemiologic molecular analysis using sequence-based typing for clinical and environmental samples.
Legionellosis Cases
Cases of pneumonia reported in Alcoi during the incubation period were considered suspected cases of legionellosis unless this diagnosis could be ruled out. A confirmed case of legionellosis was defined as a case of pneumonia with laboratory evidence of acute infection with L. pneumophila including 1) isolation of L. pneumophila serogroup 1 from respiratory secretions or lung tissue, 2) a >4-fold rise in antibody titers (from 128) against L. pneumophila serogroup1 by immunofluorescence in paired acute- and convalescent-phase serum specimens, or 3) detection of L. pneumophila serogroup 1 antigen in urine.
Cases that conformed to the case definition and/or had positive results in at least 1 of the following tests were considered suspected cases: 1) high (>256) antibody titer against L. pneumophila serogroup 1 in convalescent-phase serum, 2) seroconversion to L. pneumophila serogroup 1 by indirect inmunofluorescence in acute- and convalescent-phase serum, or 3) direct staining of bacteria in respiratory secretions or lung tissue by direct fluorescence using monoclonal or polyclonal antibodies against any Legionella spp. or serogroup, including serogroup 1.
Epidemiologic Investigation
A questionnaire was used to obtain personal information (age, gender, job, and free-time activities), clinical features (signs and symptoms, date of illness onset, date of hospitalization, previous hospitalizations, and medical history), predisposing and risk factors, place of residence, and recent urban mobility within Alcoi. Information about the patients’ addresses and usual itineraries (roads and places visited) in Alcoi was used to delimit an area of influence, or a buffer, for the location of the likely source of infection. A 500-m radius was considered around home and city itineraries for each patient by using vectorial cartography (scale 1:10,000), ortophotography (scale 1:5,000), and ArcView software (www.esri.com). The buffer for each patient was thus represented by a polygonal area; intersecting areas yielded a common area representing a likely location for the source of the outbreak.
Environmental Investigation
Systematic environmental investigations are regularly performed in the municipal water distribution system, but when this outbreak was detected, an active search for L. pneumophila was made in patients’ homes (bulk water and biofilms from showerheads and taps) and the water distribution system (bulk water); results were negative. None of the other usual sources (e.g., public fountains, cooling towers, humidifiers) were found to pose a risk. The absence of usual risk sources led us to consider other possible sources of aerosols, including moving devices used in street cleaning and asphalt repaving; the latter had been observed in the risk area during the epidemiologic inspection.
Laboratory Methods
Clinical Sampling
Respiratory samples and, when available, corresponding cultures were obtained from 11 patients with a diagnosis of legionellosis made by positive urine test result at the Hospital Virgen de los Lirios. DNA was extracted from respiratory isolates by using an UltraClean BloodSpin Kit (Mobio Laboratories, Inc., Carlsbad, CA, USA). DNA from positive L. pneumophila cultures was extracted as described (23). Briefly, bacterial colonies from pure cultures were resuspended in 200 μL of 20% Chelex 100 resin (Bio-Rad Laboratories, Richmond, CA, USA). DNA was then extracted during 3 freeze–thaw cycles (–75°C for 10 min and 94°C for 10 min), and cellular debris was removed by centrifuging at 10,000 × g for 1 min. The amount of genomic DNA was measured by spectrophotometry at 260 nm in triplicate, and DNA purity was checked by using the A260/A280 ratio. Purified DNA was stored at –20°C until used. DNA extraction from respiratory samples (sputum, bronchoalveolar aspirated secretions, and lung biopsy tissue) was performed with a QIAamp DNA Mini Kit (QIAGEN, Valencia, CA, USA) and stored at –20°C.
Environmental Sampling
Water samples were filtered and treated with acid. Water and swab specimens were plated onto buffered charcoal yeast extract medium with and without supplemental antimicrobial drugs; standard plating techniques were used. Inoculated plates were incubated at 35°C and examined regularly for colonies resembling Legionella spp. Suspected colonies were inoculated onto biplates containing buffered charcoal yeast extract medium with and without
PCR, Sequencing, and Allelic Profile Assignment
For cultured isolates, the 7 loci in the European Working Group for Legionella Infections (EWGLI, www.ewgli.org) typing scheme were amplified as detailed by Gaia et al. (17) and Ratzow et al. (18). DNA from respiratory samples was amplified by using a seminested approach. For the first PCR we used the same primers and conditions as for cultured isolates. For the second PCR we used the internal primers described by Coscollá et al. (22) and used 2 μL of the first PCR products as template. Amplification conditions and profiles were the same as for the first PCR except for the annealing temperature (22). PCR products were purified by using a High Pure PCR Product Purification Kit (Roche Diagnostics, Mannheim, Germany). PCRs were conducted in a reaction volume of 50 μL containing 20 ng of genomic DNA, 1 U of Taq DNA polymerase, 200 μmol/L of each dNTP, 2.5 mmol/L of magnesium-free buffer, 2.5 mmol/L MgCl2, and 0.2 μmol/L of each pair of primers. We adopted the gene notation used in the first L. pneumophila genome published (24); consequently, locus fliC corresponds to locus flaA used in the EWGLI typing scheme (17).
Purified DNA was directly sequenced by the dideoxy method by using a BigDye Terminator v3.0 Ready Reaction Cycle Sequencing Kit and was analyzed in an ABI PRISM 3700 sequencer (each from Applied Biosystems, Foster City, CA, USA). Sequencing of PCR products from respiratory samples differed only in the use of the same internal primers used in the second PCR of the seminested approach. Sequence chromatogram files were analyzed by using the Staden sequence analysis package (25).
Allelic profiles for the 7 sequenced genes were obtained from EWGLI and were aligned and compared with sequences derived in this study. Multiple sequence alignments were obtained by using ClustalX (26) and further refined by visual inspection.
Molecular Phylogenetic Analysis
A phylogenetic reconstruction was obtained with the concatenated alignment of sequences from the 7 loci analyzed. Models of nucleotide substitution were assessed by using the maximum-likelihood approach implemented in jModeltest (27). Maximum-likelihood phylogenetic trees were obtained with PHYML 3.0 (28) by using the previously derived models of nucleotide substitution for each locus. Support for the nodes was evaluated by bootstrapping with 1,000 pseudoreplicates.
Epidemiologic Findings
Among patients with positive urine antigen test results, 11 cases of legionellosis were confirmed and L. pneumophila was isolated from 4. All patients required hospitalization, and all except 1 recovered. (The patient who did not recover had severe signs and symptoms and subsequently died.) The main signs and symptoms were fever (100% incidence), pneumonia (100%), headache (27.3%), myalgia (27.3%), diarrhea and/or vomiting (18.2%), and confusion (45.5%). The average age was 70 years, range 49–88 years. More men than women were affected (male:female ratio = 4.5).
Confirmed cases occurred from July 21 through September 17. According to the date of disease onset, the outbreak showed 3 epidemic waves: 2 cases in the second half of July, 8 cases in the first half of August, and 1 case in the second half of August (Figure 1).
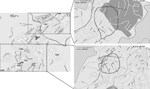
No common indoor source of exposure was found, and the initial hypothesis was that the outbreak originated from environmental contamination of an unknown source capable of producing and dispersing large quantities of aerosols contaminated with L. pneumophila. The first 2 patients lived in the northern part of the city, which suggested that the source could be located in that area. The spatial distribution of patients’ buffers changed in August, thus indicating that the likely source of the outbreak had moved to the Santa Rosa quarter (Figure 2). The area of epidemic risk was modified accordingly, and the search for putative environmental sources focused on that neighborhood.
Environmental Findings
L. pneumophila was not isolated from samples derived from the municipal water supply, traditional risk facilities, and patients’ houses. The environmental investigation was extended to other potential sources, especially portable cleaning devices such as sweepers, hydrocleaners, and water tanks used to clean the streets, all of which used water from the municipal water supply. At that time, the Santa Rosa quarter was being repaved, and 1 of the machines used in the repaving process was a tank truck that carried water used by a large milling machine. The water in the tank was obtained from a natural spring untreated with chlorine or anything else. Because the average daytime temperature in Alcoi during July and August is 27°C, the water in the machine might have been warm enough for L. pneumophila growth. The milling machine had been working north of Alcoi around July 15 and in the Santa Rosa neighborhood from July 31 through August 20 (Figure 2). This activity fits spatially and temporally with the incubation period of confirmed cases. This machine was identified and removed from service on August 21, thus was able to cause the last infection detected on August 23 (Figure 2).
Microbiological Findings
L. pneumophila serogroup 1 was isolated from the water in the tank and from the atomizers in the milling machine (Figure 3). L. pneumophila was also isolated from other machines used for street cleaning in the city, but these organisms were from serogroups other than serogroup1.
The milling machine and the water tank were immediately confined outside the city, sealed, cleaned, and disinfected. When the machine was put back into use, a new cleaning and maintenance protocol was implemented to prevent future contamination. The cleaning protocol consisted of treatment with chlorine (20 ppm) and removal of the atomizers and their replacement by gravity-based water distributors. Additionally, for operation in the city, use of a separate thermo-insulated water tank in which chlorine was continuously applied at 20 ppm was required, thus preventing any further growth of L. pneumophila. Additionally, water had to be obtained from the municipal system.
Molecular Characteristics
L. pneumophila sequence information was obtained from 7 uncultured sputum samples and from 4 cultures derived from the 11 patients studied. Complete allelic profiles, according to the EWGLI typing scheme, were obtained for 9 clinical samples in the study. All had the same profile, which corresponded to sequence type (ST) 578 in the EWGLI database (Table 1). Partial allelic profiles for the other 2 samples were consistent with ST578. The 9 L. pneumophila isolates derived from putative environmental sources, including the milling machine and the water tank (Table 2), were also characterized and corresponded to 4 STs: ST578 in 4 samples, ST1 in 3 samples, ST777 in 1 sample, and a new ST in sample 4159.
A phylogenetic tree was obtained from the concatenated alignment of the 2,984 bp (Figure 4). The phylogenetic analysis showed that 4 environmental samples were identical to the clinical samples (represented by ST578 C/E in Figure 4). Their comparison with the remaining environmental samples showed 14–50 nt differences to strains 4159 and 4143/4160/7970 (represented by ST1 in Figure 4), respectively. The comparison of these to reference strains showed that outbreak samples were more closely related to the Corby strain (29), a virulent human isolate from which it differed by only 6 nt.
During the epidemic outbreak of legionellosis in the summer of 2009 in Alcoi, Spain, 11 affected persons were identified, and their L. pneumophila isolates shared the same sequence-based typing profile (ST578). Spatial-temporal analysis of outbreak cases pointed to a milling machine used in street asphalt repaving and its water tank as the most likely source of infection. Molecular typing confirmed that L. pneumophila isolated from the machine showed the same allelic profile as the samples from the patients. When this machine was removed from service and cleaned, infections in this locality ceased.
The absence of apparent risk facilities in the area during the outbreak period and the changing spatial distribution of cases led us to consider alternative sources of contamination and spread. The heterogeneous spatial grouping led us to hypothesize that the transmission sources could be mobile. The analysis of the water tank and milling machine used in both neighborhoods where risk areas were identified during the incubation period of confirmed cases resulted in isolation of L. pneumophila. Molecular results confirmed that 4 of the 9 environmental isolates obtained showed a sequence-based typing profile identical to that of all clinical samples. This result highlights the fact that a device not previously considered to represent a risk for L. pneumophila infection, such as a street paving machine, can be associated with a legionellosis outbreak.
These kinds of machines are good candidates for spreading L. pneumophila infections, given their ability to generate aerosols. These machines are used in urban areas where contaminated aerosols can be inhaled by many citizens. They are continually moving, making their identification as a source of infection more difficult because by the time an outbreak is detected and the causative L. pneumophila strains are characterized, these machines have usually moved to another location. Lack of suitable cleaning routines for these machines makes them excellent candidates for colonization with L. pneumophila. In this particular outbreak, use of untreated water from a natural spring contributed to the contamination of the tank and milling machine. These devices are frequently stored and operated in and from industrial areas where nontreated water supplies are common, which increases risk for colonization by L. pneumophila. Whether they spread L. pneumophila more easily than other devices usually linked to such outbreaks, like cooling towers or spas, is unknown, but they should be considered risk devices for the reasons detailed above.
During 1999–2005, ST578 has been found in clinical samples from patients with legionellosis in Alcoi (22) and has caused recurrent outbreaks and sporadic cases of community-acquired pneumonia. However, to our knowledge, no identical isolate has been found in the environment in this area (M. Coscollá et al., unpub. data). Finding ST578 in a sports club (Table 2), not related to the outbreak, indicates that it can occasionally be found in other risk facilities, which would help to explain its association with past clinical cases in Alcoi. Additionally, this profile was reportedly found in an environmental strain detected in Mexico during an epidemiologic investigation of travel-associated legionellosis (EWGLI sequence-based typing database, www.ewgli.org).
Although culture isolates were available for only 4 of the 11 outbreak patients identified, our use of a sequence-based typing approach to analyze clinical samples on the basis of direct extraction and sequencing of L. pneumophila from sputum samples increased the number of patients we were able to study (i.e., all those identified in the outbreak). The efficiency of this approach has been demonstrated to be higher than that of sequencing after isolating L. pneumophila from cultures (22). Moreover, the 100% match between the sequences directly obtained from respiratory samples and cultured isolates from the same patient corroborates the suitability of the direct sequencing approach for the identification and molecular epidemiology studies of L. pneumophila (22). We think that this approach extends the usefulness of molecular epidemiologic tools in the study of L. pneumophila outbreaks, thus enabling more precise identification of their sources.
Dr Coscollá is a postdoctoral researcher in the Tuberculosis Research Unit of the Swiss Tropical and Public Health Institute, Basel, Switzerland. Her research interests are focused on pathogens’ evolution and molecular epidemiology of infectious diseases by using population genetics/genomics and molecular systematic tools.
Acknowledgments
We thank the following persons and institutions for providing samples and information: Vicente Catalán, Servicios de Laboratorio, Medicina Interna y Urgencias del Hospital Virgen de los Lirios, Laboratorio de Salud Pública de Alicante, Unidades de Epidemiología y de Sanidad Ambiental del Centro de Salud Pública de Alcoi, and Direccion General de Salud Publica–Conselleria de Sanidad.
This research was supported by DGSP-Conselleria de Sanitat de la Generalitat Valenciana (Spain) and projects BFU2008-03000 from Ministerio de Ciencia e Innovación and ACOMP/2009/240 and ACOMP/2010/148 from Conselleria d’Educació (Generalitat Valenciana).
References
- McDade JE, Shepard CC, Fraser DW, Tsai TR, Redus MA, Dowdle WR. Legionnaires’ disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N Engl J Med. 1977;297:1197–203. DOIPubMedGoogle Scholar
- Fraser DW, Tsai TR, Orenstein W, Parkin WE, Beecham HJ, Sharrar RG, Legionnaires’ disease: description of an epidemic of pneumonia. N Engl J Med. 1977;297:1189–97. DOIPubMedGoogle Scholar
- Marston BJ, Plouffe JF, File TM Jr, Hackman BA, Salstrom SJ, Lipman HB, Incidence of community-acquired pneumonia requiring hospitalization. Results of a population-based active surveillance study in Ohio. The Community-Based Pneumonia Incidence Study Group. Arch Intern Med. 1997;157:1709–18. DOIPubMedGoogle Scholar
- Falco V, Fernandez de Sevilla T, Alegre J, Ferrer A, Martinez Vazquez JM. Legionella pneumophila. A cause of severe community-acquired pneumonia. Chest. 1991;100:1007–11. DOIPubMedGoogle Scholar
- Macfarlane JT, Ward MJ, Finch RG, Macrae AD. Hospital study of adult community-acquired pneumonia. Lancet. 1982;2:255–8. DOIPubMedGoogle Scholar
- Bates JH, Campbell GD, Barron AL, McCracken GA, Morgan PN, Moses EB, Microbial etiology of acute pneumonia in hospitalized patients. Chest. 1992;101:1005–12. DOIPubMedGoogle Scholar
- Sopena N, Sabria-Leal M, Pedro-Botet ML, Padilla E, Dominguez J, Morera J, Comparative study of the clinical presentation of Legionella pneumonia and other community-acquired pneumonias. Chest. 1998;113:1195–200. DOIPubMedGoogle Scholar
- Torres A, Serra-Batlles J, Ferrer A, Jimenez P, Celis R, Cobo E, Severe community-acquired pneumonia. Epidemiology and prognostic factors. Am Rev Respir Dis. 1991;144:312–8.PubMedGoogle Scholar
- Schoonmaker D, Heimberger T, Birkhead G. Comparison of ribotyping and restriction enzyme analysis using pulsed-field gel electrophoresis for distinguishing Legionella pneumophila isolates obtained during a nosocomial outbreak. J Clin Microbiol. 1992;30:1491–8.PubMedGoogle Scholar
- Boshuizen HC, Neppelenbroek SE, van Vliet H, Schellekens JF, Boer JW, Peeters MF, Subclinical Legionella infection in workers near the source of a large outbreak of Legionnaires’ disease. J Infect Dis. 2001;184:515–8. DOIPubMedGoogle Scholar
- Keller DW, Hajjeh R, DeMaria A, Fields BS, Pruckler J, Benson RS, Community outbreak of Legionnaires’ disease: an investigation confirming the potential for cooling towers to transmit Legionella species. Clin Infect Dis. 1996;22:257–61. DOIPubMedGoogle Scholar
- Garcia-Fulgueiras A, Navarro C, Fenoll D, Garcia J, Gonzales-Diego P, Jimenez-Bunuelas T, Legionnaires’ disease outbreak in Murcia, Spain. Emerg Infect Dis. 2003;9:915–21.PubMedGoogle Scholar
- Cordes LG, Wiesenthal AM, Gorman GW, Phair JP, Sommers HM, Brown A, Isolation of Legionella pneumophila from hospital shower heads. [PMID: 7469211]. Ann Intern Med. 1981;94:195–7.PubMedGoogle Scholar
- Hlady WG, Mullen RC, Mintz CS, Shelton BG, Hopkins RS, Daikos GL. Outbreak of Legionnaire’s disease linked to a decorative fountain by molecular epidemiology. Am J Epidemiol. 1993;138:555–62.PubMedGoogle Scholar
- O’Loughlin RE, Kightlinger L, Werpy MC, Brown E, Stevens V, Hepper C, Restaurant outbreak of Legionnaires’ disease associated with a decorative fountain: an environmental and case-control study. BMC Infect Dis. 2007;7:93. DOIPubMedGoogle Scholar
- Piso RJ, Caruso A, Nebiker M. Hose as a source of Legionella pneumonia. A new risk factor for gardeners? J Hosp Infect. 2007;67:396–7. DOIPubMedGoogle Scholar
- Gaia V, Fry NK, Afshar B, Luck PC, Meugnier H, Etienne J, Consensus sequence-based scheme for epidemiological typing of clinical and environmental isolates of Legionella pneumophila. J Clin Microbiol. 2005;43:2047–52. DOIPubMedGoogle Scholar
- Ratzow S, Gaia V, Helbig JH, Fry NK, Luck PC. Addition of neuA, the gene encoding N-acylneuraminate cytidylyl transferase, increases the discriminatory ability of the consensus sequence-based scheme for typing Legionella pneumophila serogroup 1 strains. J Clin Microbiol. 2007;45:1965–8. DOIPubMedGoogle Scholar
- Hussong D, Colwell RR, O’Brien M, Weiss E, Pearson AD, Weiner RM, Viable Legionella pneumophila not detectable by culture on agar media. Nat Biotechnol. 1987;5:947–50. DOIGoogle Scholar
- Lindsay DS, Abraham WH, Findlay W, Christie P, Johnston F, Edwards GF. Laboratory diagnosis of Legionnaires’ disease due to Legionella pneumophila serogroup 1: comparison of phenotypic and genotypic methods. J Med Microbiol. 2004;53:183–7. DOIPubMedGoogle Scholar
- Schurmann D. Prevalence and diagnosis of Legionella pneumonia: a 3-year prospective study with emphasis on application of urinary antigen detection. J Infect Dis. 1990;162:1341–8. DOIPubMedGoogle Scholar
- Coscollá M, González-Candelas F. Direct sequencing of Legionella pneumophila from respiratory samples for sequence-based typing analysis. J Clin Microbiol. 2009;47:2901–5. DOIPubMedGoogle Scholar
- Coscollá M, González-Candelas F. Population structure and recombination in environmental isolates of Legionella pneumophila. Environ Microbiol. 2007;9:643–56. DOIPubMedGoogle Scholar
- Chien M, Morozova I, Shi S, Sheng H, Chen J, Gomez SM, The genomic sequence of the accidental pathogen Legionella pneumophila. Science. 2004;305:1966–8. DOIPubMedGoogle Scholar
- Staden R. The Staden sequence analysis package. Mol Biotechnol. 1996;5:233–41. DOIPubMedGoogle Scholar
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. DOIPubMedGoogle Scholar
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–6. DOIPubMedGoogle Scholar
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. DOIPubMedGoogle Scholar
- Glockner G, Albert-Weissenberger C, Weinmann E, Jacobi S, Schunder E, Steinert M, Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. Int J Med Microbiol. 2008;298:411–28. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 16, Number 9—September 2010
| EID Search Options |
|---|
|
|
|
|
|
|


Please use the form below to submit correspondence to the authors or contact them at the following address:
Mireia Coscollá, Tuberculosis Research Unit, Swiss Tropical and Public Health Institute, Socinstrasse 57, 4002 Basel, Switzerland
Top