Volume 17, Number 1—January 2011
Dispatch
Possible Interruption of Measles Virus Transmission, Uganda, 2006–2009
Figure 2
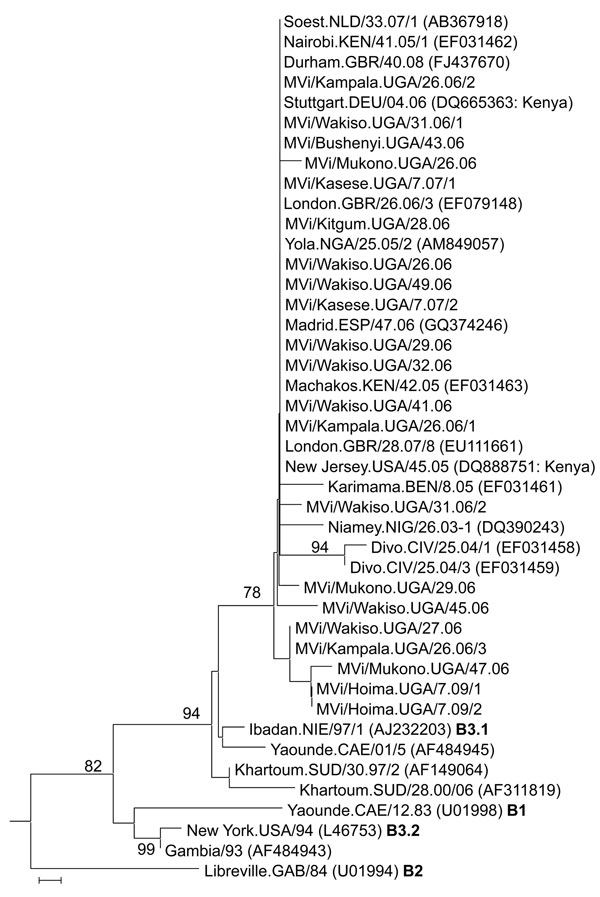
Figure 2. Phylogenetic analysis of the relationship between sequences of 21 Ugandan measles virus isolates obtained during 2006–2009 and 22 other recently described clade B nucleoprotein (N) gene sequences, including the World Health Organization reference strains for the B clade (13). Boldface indicates different genotypes. Analyses are based on sequences of the 450 nt encoding the COOH-terminal 150 nt of the N gene. The unrooted neighbor-joining consensus tree was generated by bootstrap analysis of 500 replicates by using MEGA4 software (www.megasoftware.net). Bootstrap percentages are shown when >75%. Only names of the isolates from Uganda (UGA) start with “MVi,” and all comparison strains have their GenBank accession numbers indicated in parentheses. Genotypes of the World Health Organization reference sequences are indicated after the accession number. Comparison sequences were from viruses isolated in Benin (BEN), Cameroon (CAE), Côte d’Ivoire (CIV), Gabon (GAB), the Gambia, Germany (DEU), Great Britain (GBR), Kenya (KEN), Niger (NIG), Nigeria (NIE/NGA), Spain (ESP), Sudan (SUD), the Netherlands (NLD), and the United States (USA). Sequences from Uganda were most closely related to the B3.1 viruses of the September–December 2005 measles outbreak in Kenya (8). Scale bar indicates nucleotide substitutions per site.
References
- Mbabazi WB, Nanyunja M, Makumbi I, Braka F, Baliraine FN, Kisakye A, Achieving measles control: lessons from the 2002–06 measles control strategy for Uganda. Health Policy Plan. 2009;24:261–9. DOIPubMedGoogle Scholar
- Rota PA, Featherstone DA, Bellini WJ. Molecular epidemiology of measles virus. In: Griffin DE, Oldstone MBA, editors. Measles: pathogenesis and control. Berlin: Springer-Verlag; 2009. p. 129–50.
- Muwonge A, Nanyunja M, Rota PA, Bwogi J, Lowe L, Liffick SL, New measles genotype, Uganda. Emerg Infect Dis. 2005;11:1522–6.PubMedGoogle Scholar
- World Health Organization. Manual for the laboratory diagnosis of measles and rubella virus infection. 2nd ed. Geneva (Switzerland); The Organization; 2007.
- Smit SB, Hardie D, Tiemessen CT. Measles virus genotype B2 is not inactive: evidence of continued circulation in Africa. J Med Virol. 2005;77:550–7. DOIPubMedGoogle Scholar
- Kouomou DW, Nerrienet E, Mfoupouendoun J, Tene G, Whittle H, Wild TF. Measles virus strains circulating in central and west Africa: geographical distribution of two B3 genotypes. J Med Virol. 2002;68:433–40. DOIPubMedGoogle Scholar
- El Mubarak HS, van de Bildt MW, Mustafa OA, Vos HW, Mukhtar MM, Ibrahim SA, Genetic characterization of wild-type measles viruses circulating in suburban Khartoum, 1997–2000. J Gen Virol. 2002;83:1437–43.PubMedGoogle Scholar
- Rota J, Lowe L, Rota P, Bellini W, Redd S, Dayan G, Identical genotype B3 sequences from measles patients in 4 countries, 2005. Emerg Infect Dis. 2006;12:1779–81.PubMedGoogle Scholar
- Central Intelligence Agency. The world fact book: Uganda [cited 2010 Aug 18]. https://www.cia.gov/library/publications/the-world-factbook/geos/ug.html
- Ferrari MJ, Grais RF, Bharti N, Conlan AJ, Bjornstad ON, Wolfson LJ, The dynamics of measles in sub-Saharan Africa. Nature. 2008;451:679–84. DOIPubMedGoogle Scholar
- World Health Organization. Implementing intregrated disease surveillance and response. Wkly Epidemiol Rec. 2003;78:233–40.
- World Health Organization. The RED strategy [cited 2010 May 13]. http://www.who.int/immunization_delivery/systems_policy/red/en
- World Health Organization. New genotype of measles virus and update on global distribution of measles genotypes. Wkly Epidemiol Rec. 2005;80:348–51.
- Biellik R, Madema S, Taole A, Kutsulukuta A, Allies E, Eggers R, First 5 years of measles elimination in southern Africa: 1996–2000. Lancet. 2002;359:1564–8. DOIPubMedGoogle Scholar
- Otten MW Jr, Okwo-Bele JM, Kezaala R, Biellik R, Eggers R, Nshimirimana D. Impact of alternative approaches to accelerated measles control: experience in the African region, 1996–2002. J Infect Dis. 2003;187:S36–43. DOIPubMedGoogle Scholar