Volume 17, Number 11—November 2011
THEME ISSUE
CHOLERA IN HAITI
Research
Comparative Genomics of Vibrio cholerae from Haiti, Asia, and Africa
Figure 1
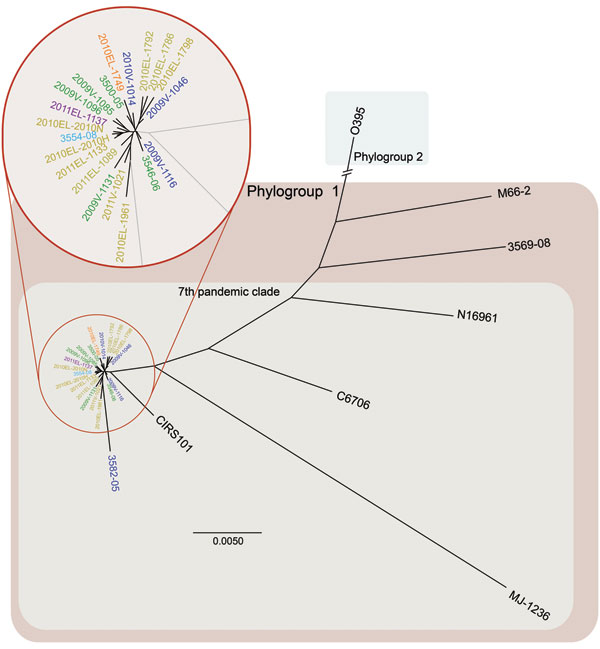
Figure 1. Whole-genome neighbor-joining tree of Vibrio cholerae isolate from cholera outbreak in Haiti, fall 2010; concurrent clinical isolates with pulsed-field gel electrophoresis pattern-matched combinations; reference isolates sequenced in this study; and available reference sequences. Sequence alignments of quality draft or complete genomes were performed by using Progressive Mauve (16) and visualized by using PhyML version 3.0 (17). Whole-genome relationship of Haiti isolates with closest genetic relatives is shown in the inset. Scale bar indicates nucleotides substitutions per site.
References
- Feng L, Reeves PR, Lan R, Ren Y, Gao C, Zhou Z, A recalibrated molecular clock and independent origins for the cholera pandemic clones. PLoS ONE. 2008;3:e4053. DOIPubMedGoogle Scholar
- Safa A, Nair GB, Kong RY. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol. 2010;18:46–54. DOIPubMedGoogle Scholar
- Nguyen BM, Lee JH, Cuong NT, Choi SY, Hien NT, Anh DD, Cholera outbreaks caused by an altered Vibrio cholerae O1 El Tor biotype strain producing classical cholera toxin B in Vietnam in 2007 to 2008. J Clin Microbiol. 2009;47:1568–71. DOIPubMedGoogle Scholar
- Grim CJ, Hasan NA, Taviani E, Haley B, Chun J, Brettin TS, Genome sequence of hybrid Vibrio cholerae O1 MJ-1236, B-33, and CIRS101 and comparative genomics with V. cholerae. J Bacteriol. 2010;192:3524–33. DOIPubMedGoogle Scholar
- Pan American Health Organization. Cholera and post-earthquake response in Haiti, 2011 Jul 25 [cited 2011 Aug 6]. http://www.who.int/hac/crises/hti/sitreps/haiti_health_cluster_bulletin_25july2011.pdf.
- Centers for Disease Control and Prevention. Update on cholera—Haiti, Dominican Republic, and Florida, 2010. MMWR Morb Mortal Wkly Rep. 2010;59:1637–41.PubMedGoogle Scholar
- Gilmour MW, Martel-Laferrière V, Lévesque S, Gaudreau C, Bekal S, Nadon C, Vibrio cholerae in traveler from Haiti to Canada. Emerg Infect Dis. 2011;17:1124–5.PubMedGoogle Scholar
- Talkington D, Bopp C, Tarr C, Parsons MB, Dahourou G, Freeman M, Characterization of toxigenic Vibrio cholerae from Haiti, 2010–2011. Emerg Infect Dis. 2011;17:2122–9.
- Chin CS, Sorenson J, Harris JB, Robins WP, Charles RC, Jean-Charles RR, The origin of the Haitian cholera outbreak strain. N Engl J Med. 2011;364:33–42. DOIPubMedGoogle Scholar
- Archer BN, Cengimbo A, De Jong GM, Keddy KH, Smith AM, Sooka A, Cholera outbreak in South Africa: preliminary descriptive epidemiology on laboratory-confirmed cases, 15 November 2008 to 30 April 2009. Communicable Diseases Surveillance Bulletin. 2009;7:3–8.
- Sakazaki R, Shimada T. Serovars of Vibrio cholerae identified during 1970–1975. Jpn J Med Sci Biol. 1977;30:279–82.PubMedGoogle Scholar
- Sakazaki R, Tamura K, Gomez CZ, Sen R. Serological studies on the cholera group of vibrios. Jpn J Med Sci Biol. 1970;23:13–20.PubMedGoogle Scholar
- Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–83. DOIPubMedGoogle Scholar
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–9. DOIPubMedGoogle Scholar
- Meyer F, Goesmann A, McHardy AC, Bartels D, Bekel T, Clausen J, GenDB–an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 2003;31:2187–95. DOIPubMedGoogle Scholar
- Darling AE, Mau B, Perna NT. Progressive Mauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5:e11147. DOIPubMedGoogle Scholar
- Guindon S, Delsuc F, Dufayard JF, Gascuel O. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol. 2009;537:113–37. DOIPubMedGoogle Scholar
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. DOIPubMedGoogle Scholar
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–52. DOIPubMedGoogle Scholar
- Petkau A, Stuart-Edwards M, Stothard P, Van Domselaar G. Interactive microbial genome visualization with GView. Bioinformatics. 2010;26:3125–6. DOIPubMedGoogle Scholar
- Mason PR. Zimbabwe experiences the worst epidemic of cholera in Africa. J Infect Dev Ctries. 2009;3:148–51. DOIPubMedGoogle Scholar
- Chun J, Grim CJ, Hasan NA, Je HL, Seon YC, Haley BJ, Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A. 2009;106:15442–7. DOIPubMedGoogle Scholar
- Olsvik O, Wahlberg J, Petterson B, Uhlen M, Popovic T, Wachsmuth IK, Use of automated sequencing of polymerase chain reaction–generated amplicons to identify three types of cholera toxin subunit B in Vibrio cholerae O1 strains. J Clin Microbiol. 1993;31:22–5.PubMedGoogle Scholar
- Gilmour MW, Graham M, Van Domselaar G, Tyler S, Kent H, Trout-Yakel KM, High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics. 2010;11:120.PubMedGoogle Scholar
- Gardy JL, Johnston JC, Ho Sui SJ, Cook VJ, Shah L, Brodkin E, Whole genome sequencing and social network analysis of a tuberculosis outbreak. N Engl J Med. 2011;364:730–9. DOIPubMedGoogle Scholar
- Taboada EN, van Belkum A, Yuki N, Acedillo RR, Godschalk PC, Koga M, Comparative genomic analysis of Campylobacter jejuni associated with Guillain-Barré and Miller Fisher syndromes: neuropathogenic and enteritis-associated isolates can share high levels of genomic similarity. BMC Genomics. 2007;8:359. DOIPubMedGoogle Scholar
- Cho YJ, Yi H, Lee JH, Kim DW, Chun J. Genomic evolution of Vibrio cholerae. Curr Opin Microbiol. 2010;13:646–51. DOIPubMedGoogle Scholar
- Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–4. DOIPubMedGoogle Scholar
- Goel AK, Jain M, Kumar P, Bhadauria S, Kmboj DV, Singh L. A new variant of Vibrio cholerae O1 El Tor causing cholera in India. J Infect. 2008;57:280–1. DOIPubMedGoogle Scholar
- Quilici ML, Massenet D, Gake B, Bwalki B, Olson DM. Vibrio cholerae O1 variant with reduced susceptibility to ciprofloxacin, western Africa. Emerg Infect Dis. 2010;16:1804–5.PubMedGoogle Scholar
1Members of the V. cholerae Outbreak Genomics Task Force are Arunmozhi Balajee, Shanna Bolcen, Cheryl A. Bopp, John Besser, Ifeoma Ezeoke, Patricia Fields, Molly Freeman, Lori Gladney, Dhwani Govil, Michael S. Humphrys, Maria Sjölund-Karlsson, Karen H. Keddy, Elizabeth Neuhaus, Michele M. Parsons, Efrain Ribot, Maryann Turnsek, Shaun Tyler, Jean M. Whichard, Anne Whitney, and the authors.