Volume 17, Number 3—March 2011
Research
Nontuberculous Mycobacteria from Household Plumbing of Patients with Nontuberculous Mycobacteria Disease
Cite This Article
Citation for Media
Abstract
To determine whether plumbing could be a source of nontuberculous mycobacteria (NTM) infection, during 2007–2009 I isolated NTM from samples from household water systems of NTM patients. Samples from 22/37 (59%) households and 109/394 (28%) total samples yielded NTM. Seventeen (46%) of the 37 households yielded >1 Mycobacterium spp. isolate of the same species as that found in the patient; in 7 of those households, the patient isolate and 1 plumbing isolate exhibited the same repetitive sequence-based PCR DNA fingerprint. Households with water heater temperatures <125°C (<50°C) were significantly more likely to harbor NTM compared with households with hot water temperatures >130°F (>55°C) (p = 0.0107). Although households with water from public or private water systems serving multiple households were more likely to have NTM (19/27, 70%) compared with households with a well providing water to only 1 household (5/12, 42%), that difference was not significant (p = 0.1532).
Nontuberculous mycobacteria (NTM) are opportunistic pathogens found in the environment (e.g., water and soil) and cause life-threatening infections in humans, other mammals, and birds (1,2). The incidence of NTM disease in Canada and the United States seems to be increasing (3–5). In Toronto, Ontario, Canada, NTM disease incidence rose from 1.5 to 9.0 cases per 100,000 population during 1997–2003 (3). The most common NTM infecting persons in the United States are Mycobacterium avium, M. intracellulare, and M. avium complex (MAC) (6). Infections occur in immunodeficient (e.g., HIV/AIDS) and immunosuppressed (e.g., cancer and transplant) patients and nonimmunosuppressed persons with the classic risk factors for mycobacteria infection, which include exposure to dust or smoke and underlying lung disease (6,7). Cystic fibrosis (8), heterozygosity for mutations in the cystic fibrosis transmembrane conductance regulator gene (9), and α-1-antitrypsin deficiency (10) predispose persons to NTM disease. Elderly, slender women lacking any of the classic risk factors for NTM disease are also at risk for NTM pulmonary disease (11–13). The major manifestation of NTM infection in the immunocompetent host is pulmonary disease, whereas disseminated disease (i.e., bacteremia) is found in patients with AIDS and other immunosuppressed persons (6).
NTM, particularly M. avium and M. intracellulare, have been recovered from a variety of environmental niches with which humans come in contact, especially drinking water (14–19). NTM are not transient contaminants of drinking water distribution systems; rather, the NTM grow and persist in plumbing (19,20). For example, numbers of mycobacteria increase in pipes as the distance from the treatment plant increases (19). NTM cell surface hydrophobicity results in disinfectant resistance and a predilection to attach to surfaces where NTM grow and form biofilms (21,22) that further increase disinfectant resistance (23). Because disinfectants inhibit the competing microflora, the slow-growing NTM can grow on the available nutrients in the absence of competition. M. avium can grow in drinking water at concentrations of assimilable organic carbon of >50 µg/L (24). Thus, there is strong reason to hypothesize that NTM can colonize and persist in household plumbing.
Sources of human infection with NTM, including MAC, have been found in water (18) and potting soil (25). Notably, M. avium was detected in water aboard the Russian space station Mir (26). Recently, researchers found that the DNA fingerprints of several M. avium isolates recovered from the shower of an M. avium–infected patient were almost identical to isolates recovered from the patient, indicating that the household water could have been the source of the patient’s pulmonary disease (27). Despite that evidence, several publications have documented low frequency of recovery of MAC from household water samples (17,28–30). Such low recovery rates of M. avium and M. intracellulare could be because water samples, not biofilm, were collected. As MAC preferentially attaches to surfaces (21–23), MAC may be at low numbers in water samples. Furthermore, in the studies cited above, a low number (<4) of samples were collected from individual households. Recovery of multiple NTM or MAC isolates is necessary because of the clonal variation of MAC (25,27). The pilot study described here isolated, enumerated, and DNA fingerprinted NTM from households of patients with NTM to test the hypothesis that household plumbing could be a source of their NTM infection.
Patients and NTM Isolates
NTM patients were recruited to participate in studies of their household water systems through the auspices of Nontuberculous Mycobacteria Research and Information, Inc. Informed consent was obtained from each participating patient, and the study was reviewed by the Virginia Tech Institutional Research Board and granted exempt status. NTM isolates from the patients, if possible, were obtained through collaborating physicians and mycobacteriology laboratories. In some instances multiple patient isolates of different species were found. A questionnaire was provided to each patient to obtain information about the household plumbing.
Household Water and Biofilm Samples
Sterile containers and swabs were sent to each collaborating patient household. Directions for collection of hot and cold water samples (500 mL) and biofilms/sediment from water taps and showerheads by using swabs were provided. If the patient thought that infection might have occurred as a result of exposure to soil, soil samples were collected. In some cases filters (fiber, activated charcoal, and reverse osmosis) were collected. All samples were returned at ambient temperatures by express courier service to the Mycobacteriology Laboratory in the Department of Biological Sciences at Virginia Tech.
Isolation and Identification of Mycobacteria
Mycobacteria in water and swab (taps and filters) samples were counted and isolated as described (27). Soil samples were processed as described (25). Most acid-fast colonies picked for identification and enumeration were small (1-mm diameter after 14 days at 37°C), unpigmented to yellow, and resembled either the transparent or opaque types previously reported (17). Acid-fast isolates were identified by nested PCR of the 16S rRNA gene (31) and PCR amplification and analysis of restriction endonuclease digestion fragments of the heat-shock protein 65 (hsp65) gene (32).
Fingerprinting Patient and Environmental Isolates
In those instances in which the Mycobacterium species from the patient and household water system isolates were the same, isolates were fingerprinted by repetitive sequence-based PCR (rep-PCR) (33). Matches were confirmed by use of GelCompar II software (Applied Maths, Inc., Austin, TX, USA).
Household Plumbing Samples
Samples for NTM isolation were received from 31 collaborating patients throughout the United States and Canada: Arizona, California, Colorado, Connecticut, Florida, Georgia, Michigan, New Jersey, New York, Pennsylvania, Texas, Vermont, Virginia, and Wisconsin, USA; and Ontario, Canada. Six patients each had 2 residences and sent samples from each residence.
NTM Isolation
The isolates from the 31 patients with NTM infection included M. avium (9), M. intracellulare (6), MAC (11), M. abscessus (4), and M. xenopi (1). Isolates could not be obtained from 11 patients, thus preventing rep-PCR fingerprinting even in those instances where household isolates belonged to the same species. Thus, the total number of patient isolates available for fingerprinting was only 20. All putative Mycobacterium spp. isolates recovered from samples were identified, and 45% of NTM-positive households (10/22) and 1.5% of NTM-positive samples (6/394) yielded >1 NTM species (Table 1). The average number of different NTM species per household was 1.9 (range 1–5 NTM species/household). In those instances where the Mycobacterium species of the patient and their household plumbing isolates were the same (e.g., M. avium), all isolates belonging to the same species as the patient were subject to rep-PCR fingerprinting. Household isolates included M. avium (10), M. intracellulare (10), M. malmoense (5), M. szulgai (3), M. chelonae (2), M. gordonae (6), and 1 each of M. scrofulaceum, M. terrae, and M. trivale (Table 1). Samples were coded with the first 2 or 3 letters representing each patient, a letter representing sample type (W, water; Sw, swab [biofilm]; S, soil), a number for sample number from a household collection, and the final number for the isolate from the sample; thus, ML-W-6-2 is the second water isolate from the sixth sample collected from patient ML’s household.
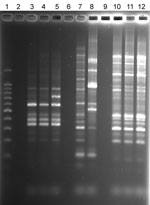
NTM were isolated from water, biofilm, filter, or soil samples from 22 (59%) households sampled and from 109 (28%) of 394 samples. There was a positive correlation between the number of samples collected per household and the number of NTM-positive samples (r = 0.4581). In 8 households >50% of the samples yielded NTM, and in 7 households no NTM were isolated. Seventeen of the 37 household sample collections had at least 1 sample that yielded an NTM isolate that belonged to the same species as that of the patient. Among those 17 households, at least 1 NTM isolate from 7 households exhibited the same rep-PCR fingerprint as that of the patient. Specifically, the Figure illustrates matching rep-PCR band patterns of patient isolate ML-P-1 (lane 3) and shower water isolate ML-W-6-2 (lane 4) from the patient’s home and patient isolate TC-P-1 (lane 10) and tap water isolate TC-W-2-2 (lane 12) from the patient’s home. Matches were confirmed by use of GelCompar II software (Applied Maths, Inc.). Furthermore, the Figure also illustrates the relative similarity in rep-PCR band patterns of patients and their household isolates and the wide differences between isolates of different patients (compare lanes 3–4, lanes 7–8, and lanes 10–12). On the basis of diversity of band patterns and the number of bands (7–14 bands), the results confirm the discriminatory power of rep-PCR fingerprinting (32). The percentage of fingerprint matches may be an underestimate because patient isolates could not be obtained for 11/31 patients, all of whom had MAC infections.
The frequency of NTM recovery from water (47/195, 24%), biofilm (46/165, 28%), filters (4/12, 33%), and soil samples (3/17, 18%) did not differ markedly. The highest numbers of NTM, as CFUs, were recovered from biofilms (10,371 CFU/cm2), with lower numbers from filters (1,987 CFU/cm2), soils (1,500 CFU/g), and water (157 CFU/mL). Most biofilm samples were collected by swabbing either the inside of a water tap or showerhead with a sterile swab that was immediately placed in 2 mL sterile tap water. Because the samples were shipped immediately after collection, there was little opportunity for the NTM numbers to change.
Household Plumbing Characteristics as Determinants of NTM Presence
Review of the responses to the NTM patient questionnaire led to identification of 2 factors that seemed to influence NTM in household samples. Households with water heater temperatures <125°C (50°C) were more likely to yield NTM (17/20, 85%) compared with households in which water temperature was >130°F (55°C) (6/15, 40%) (Table 2). That difference was significant (p = 0.0107; relative risk 2.125, by Fisher exact test). Although households with water from a public or private water system were more likely to have NTM (19/27, 70%) compared with households with water from a well (5/12, 42%) that difference was not significant (p = 0.1532; relative risk 1.689 by Fisher exact test) (Table 3).
The data document the relevance of household water as a source of NTM infection. Seven (41%) of the 17 patients from whom isolates were obtained were infected with an NTM strain having the same DNA fingerprint as at least 1 NTM isolate from their household plumbing. Several characteristics of household water and plumbing are conducive to NTM survival and growth. Specifically, residual disinfectant selects for disinfectant-resistant NTM (23), pipe surfaces offer opportunities for biofilm formation (21–23), and low organic matter content permits growth of the oligotrophic NTM (22,24).
The frequency of samples yielding NTM (28%) reported is almost identical to the frequency of Mycobacterium spp. 16S rRNA sequences in biofilm (swab) samples collected from showers across the United States (34). In as much as that culture-independent study (34) did not collect samples specifically from households of NTM patients, apparently NTM are quite frequent in household water and plumbing across the United States and Canada and are not unique to household plumbing of NTM patients. In addition to exposure, host factors (6–10) are influential factors in the acquisition of NTM disease. For the study reported here, NTM patient contamination of samples was unlikely because the patients were either free of NTM in sputum or were continuing antimycobacteria therapy; none were persistently sputum positive. The low frequency of recovery of NTM by other studies (17,28–30) was likely because a low number of samples were collected from households. As shown here, only 28% of household samples yielded NTM, and there was a positive correlation between the number of samples collected and the recovery of NTM from household samples.
In addition to documenting the presence of NTM in households across the United States, the data from this pilot study with its relatively small sample size suggest that water heater temperature and water source could be factors influencing NTM presence. NTM were less frequently recovered from household samples whose water heater temperature was >130°C (>55°C). The relative risk of NTM presence was 2.125 for households whose water heater temperature was <125°C (<50°C). In fact, 6 of the 7 households whose patient and plumbing isolates shared identical rep-PCR patterns had water heater temperatures <125°C (<50°C). That association correlates with the temperature sensitivity of NTM species. For example, the time required to kill 90% of M. avium cells is 1,000 min at 50°C but only 54 min at 55°C; similar times were measured for M. intracellulare (35). High water heater temperatures have been associated with low numbers of Legionella spp. in household and other building plumbing (36–39).
It would follow that persons infected or at risk for NTM disease, e.g., slender elderly persons or cystic fibrosis transmembrane conductance regulator gene heterozygotes (8–13), consider increasing water heater temperatures. Households whose water came from a public or private water system were more likely to have NTM in household water than those whose water source was a well (p = 0.1532, relative risk = 1.689). Although not significant, that result is consistent with the fact that NTM are seldom detected in groundwater (40). This pilot study will be followed by an investigation to assess the influence of a variety of household plumbing characteristics in households of additional NTM patients and their neighbors.
Dr Falkinham is a professor of microbiology in the Department of Biological Sciences at Virginia Polytechnic Institute and State University. His research interests include the epidemiology of Mycobacterium avium, metal oxidation and reduction in biofilms and predatory bacteria ecology.
Acknowledgments
I thank Myra D. Williams, who performed the technical work, and the NTM patients who participated in the study.
The study was supported by a grant from the Nontuberculous Mycobacteria Research and Information Foundation, Inc.
References
- Falkinham JO III. Epidemiology of infection by nontuberculous mycobacteria. Clin Microbiol Rev. 1996;9:177–215.PubMedGoogle Scholar
- Falkinham JO III. Surrounded by mycobacteria: nontuberculous mycobacteria in the human environment. J Appl Microbiol. 2009;107:356–67. DOIPubMedGoogle Scholar
- Marras TK, Chedore P, Ying AM, Jamieson F. Isolation prevalence of pulmonary non-tuberculous mycobacteria in Ontario, 1997–2003. Thorax. 2007;62:661–6. DOIPubMedGoogle Scholar
- Iseman MD, Marras TK. The importance of nontuberculous mycobacterial lung disease. Am J Respir Crit Care Med. 2008;178:999–1000. DOIPubMedGoogle Scholar
- Billinger ME, Olivier KN, Viboud C, Montes de Oca R, Steiner C, Holland SM, Nontuberculous mycobacteria–associated lung disease in hospitalized persons, United States, 1998–2005. Emerg Infect Dis. 2009;15:1562–9.PubMedGoogle Scholar
- Marras TK, Daley CL. Epidemiology of human pulmonary infection with nontuberculous mycobacteria. Clin Chest Med. 2002;23:553–67. DOIPubMedGoogle Scholar
- Wolinsky E. Nontuberculous mycobacteria and associated diseases. Am Rev Respir Dis. 1979;119:107–59.PubMedGoogle Scholar
- Olivier KN, Weber DJ, Wallace RJ Jr, Falz AR, Lee J-H, Zhang Y, Nontuberculous mycobacteria. I. Multicenter prevalence study in cystic fibrosis. Am J Respir Crit Care Med. 2003;167:828–34. DOIPubMedGoogle Scholar
- Kim JS, Tanaka N, Newell JD, De Groote MA, Fulton K, Huitt G, Nontuberculous mycobacterial infection. CT scan findings, genotype, and treatment responsiveness. Chest. 2005;128:3863–9. DOIPubMedGoogle Scholar
- Chan ED, Kaminska AM, Gill W, Chmura K, Feldman NE, Bai X, Alpha-1-antitrypsin (AAT) anomalies are associated with lung disease due to rapidly growing mycobacteria and AAT inhibits Mycobacterium abscessus infection in macrophages. Scand J Infect Dis. 2007;39:690–6. DOIPubMedGoogle Scholar
- Prince DS, Peterson DD, Steiner RM, Gottlieb JE, Scott R, Israel HL, Infection with Mycobacterium avium complex in patients without predisposing conditions. N Engl J Med. 1989;321:863–8. DOIPubMedGoogle Scholar
- Reich JM, Johnson RE. Mycobacterium avium complex pulmonary disease. Incidence, presentation, and response to therapy in a community setting. Am Rev Respir Dis. 1991;143:1381–5.PubMedGoogle Scholar
- Kennedy TP, Weber DJ. Nontuberculous mycobacteria. An underappreciated cause of geriatric lung disease. Am J Respir Crit Care Med. 1994;149:1654–8.PubMedGoogle Scholar
- Covert TC, Rodgers MR, Reyes AL, Stelma GN Jr. Occurrence of nontuberculous mycobacteria in environmental samples. Appl Environ Microbiol. 1999;65:2492–6.PubMedGoogle Scholar
- du Moulin GC, Stottmeier C, Pelletier KD, Tsang AY, Hedley-Whyte J. Concentration of Mycobacterium avium by hospital host water systems. JAMA. 1988;260:1599–601. DOIPubMedGoogle Scholar
- Fischeder R, Schulze-Röbbecke R, Weber A. Occurrence of mycobacteria in drinking water samples. Zentralbl Hyg Umweltmed. 1991;192:154–8.PubMedGoogle Scholar
- Glover N, Holtzman A, Aronson T, Froman S, Berlin OGW, Dominguez P, The isolation and identification of Mycobacterium avium complex (MAC) recovered from Los Angeles potable water, a possible source of infection in AIDS patients. Int J Environ Health Res. 1994;4:63–72. DOIGoogle Scholar
- von Reyn CF, Maslow JN, Barber TW, Falkinham JO III, Arbeit RD. Persistent colonisation of potable water as a source of Mycobacterium avium infection in AIDS. Lancet. 1994;343:1137–41. DOIPubMedGoogle Scholar
- Falkinham JO III, Norton CD, LeChevallier MW. Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other mycobacteria in drinking water distribution systems. Appl Environ Microbiol. 2001;67:1225–31. DOIPubMedGoogle Scholar
- Hilborn ED, Covert TC, Yakrus MA, Harris SI, Donnelly SF, Rice EW, Persistence of nontuberculous mycobacteria in a drinking water system after addition of filtration treatment. Appl Environ Microbiol. 2006;72:5864–9. DOIPubMedGoogle Scholar
- Torvinen E, Suomalainen S, Lehtola MJ, Miettinen IT, Zacheus O, Paulin L, Mycobacteria in water and loose deposits of drinking water distribution systems in Finland. Appl Environ Microbiol. 2004;70:1973–81. DOIPubMedGoogle Scholar
- Williams MM, Yakrus MA, Arduino MJ, Cooksey RC, Crane CB, Banerjee SN, Structural analysis of biofilm formation by rapidly and slowly growing nontuberculous mycobacteria. Appl Environ Microbiol. 2009;75:2091–8. DOIPubMedGoogle Scholar
- Steed KA, Falkinham JO III. Effect of growth in biofilms on chlorine susceptibility of Mycobacterium avium and Mycobacterium intracellulare. Appl Environ Microbiol. 2006;72:4007–11. DOIPubMedGoogle Scholar
- Norton CD, LeChevallier MW, Falkinham JO III. Survival of Mycobacterium avium in a model distribution system. Water Res. 2004;38:1457–66. DOIPubMedGoogle Scholar
- De Groote MA, Pace NR, Fulton K, Falkinham JO III. Relationships between Mycobacterium isolates from patients with pulmonary mycobacterial infection and potting soils. Appl Environ Microbiol. 2006;72:7602–6. DOIPubMedGoogle Scholar
- Kawamura Y, Li Y, Liu H, Huang X, Li Z, Ezaki T. Bacterial population in Russian space station “Mir.”. Microbiol Immunol. 2001;45:819–28.PubMedGoogle Scholar
- Falkinham JO III, Iseman MD, De Haas P, van Soolingen D. Mycobacterium avium in a shower linked to pulmonary disease. J Water Health. 2008;6:209–13.PubMedGoogle Scholar
- Yajko DM, Chin DP, Gonzalez PC, Nassos PS, Hopewell PC, Reingold AL, Mycobacterium avium complex in water, food, and soil samples collected from the environment of HIV-infected individuals. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;9:176–82.PubMedGoogle Scholar
- Peters M, Müller C, Rüsch-Gerdes S, Seidel C, Göbel U, Pohle HD, Isolation of atypical mycobacteria from tap water in hospitals and homes: is this a possible source of disseminated MAC infection in AIDS patients? J Infect. 1995;31:39–44. DOIPubMedGoogle Scholar
- Montecalvo MA, Forester F, Tsang AY, du Moulin G, Wormser GP. Colonisation of potable water with Mycobacterium avium complex in homes of HIV-infected patients. Lancet. 1994;343:1639. DOIPubMedGoogle Scholar
- Wilton S, Cousins D. Detection and identification of multiple mycobacterial pathogens by DNA amplification in a single tube. PCR Methods Appl. 1992;1:269–73.PubMedGoogle Scholar
- Telenti A, Marchesi F, Balz M, Bally F, Böttger EC, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993;31:175–8.PubMedGoogle Scholar
- Cangelosi GA, Freeman RJ, Lewis KN, Livingston-Rosanoff D, Shah KS, Milan SJ, Evaluation of high-throughput repetitive sequence-based PCR system for DNA fingerprinting of Mycobacterium tuberculosis and Mycobacterium avium complex strains. J Clin Microbiol. 2004;42:2685–93. DOIPubMedGoogle Scholar
- Feazel LM, Baumgartner LK, Peterson KL, Frank DN, Harris JK, Pace NR. Opportunistic pathogens enriched in showerhead biofilms. Proc Natl Acad Sci U S A. 2009;106:16393–9. DOIPubMedGoogle Scholar
- Schulze-Röbbecke R, Buchholtz K. Heat susceptibility of aquatic mycobacteria. Appl Environ Microbiol. 1992;58:1869–73.PubMedGoogle Scholar
- Alary M, Joly JR. Risk factors for contamination of domestic hot water systems by legionellae. Appl Environ Microbiol. 1991;57:2360–7.PubMedGoogle Scholar
- Dewailly E, Jolly JR. Contamination of domestic water heaters with Legionella pneumophila: impact of water temperature on growth and dissemination of the bacterium. Environ Toxicol Water Qual. 1991;6:249–57. DOIGoogle Scholar
- Straus WL, Plouffe JF, File TM Jr, Lipman HB, Hackman BH, Salstrom S-J, Risk factors for domestic acquisition of Legionnaires disease. Arch Intern Med. 1996;156:1685–92. DOIPubMedGoogle Scholar
- Martinelli F, Caruso A, Moschini L, Turano A, Scarcella C, Speziani F. A comparison of Legionella pneumophila occurrence in hot water tanks and instantaneous devices in domestic, nosocomial, and community environments. Curr Microbiol. 2000;41:374–6. DOIPubMedGoogle Scholar
- Martin EC, Parker BC, Falkinham JO III. Epidemiology of infection by nontuberculous mycobacteria. VII. Absence of mycobacteria in southern groundwaters. Am Rev Respir Dis. 1987;136:344–8.PubMedGoogle Scholar
Figure
Tables
Cite This ArticleTable of Contents – Volume 17, Number 3—March 2011
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Joseph O. Falkinham, III, Department of Biological Sciences, Virginia Polytechnic Institute and State University, Blacksburg, VA 24061-0406, USA
Top