Volume 17, Number 8—August 2011
Research
Human Polyomavirus Related to African Green Monkey Lymphotropic Polyomavirus
Figure 2
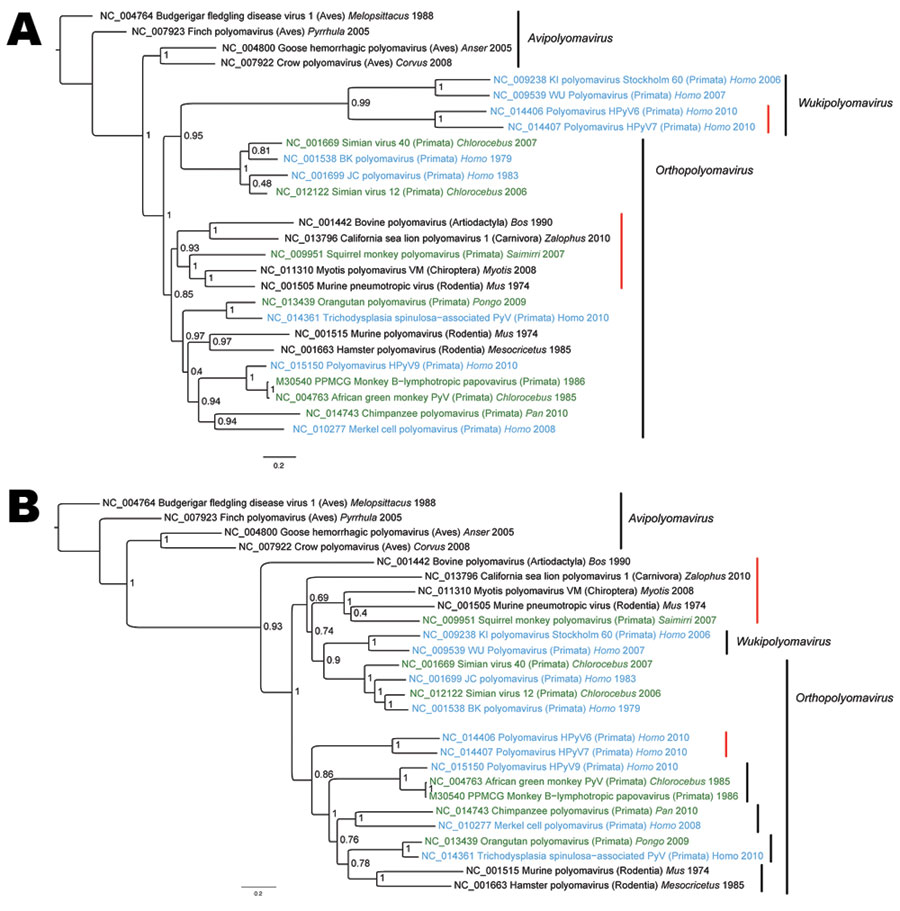
Figure 2. A) Viral protein 1 (VP1) and B) large T antigen (LT) nucleotide-based phylogenetic reconstructions of polyomaviruises inferred by using a Bayesian method. Taxa annotations include reference number, name of the virus, host taxonomic order (in parentheses), host genus whenever available, and reported collection date. Human viruses are indicated in blue, and monkey viruses are indicated in green. Red vertical bars highlight groups for which VP1 and LT signals are incongruent. Posterior probabilities are indicated at each node. GenBank identification numbers are indicated directly on trees for each sequence. Scale bars indicate nucleotide substitutions per site.
Page created: August 16, 2011
Page updated: August 16, 2011
Page reviewed: August 16, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.