Subclinical Avian Influenza A(H5N1) Virus Infection in Human, Vietnam
Mai Quynh Le

, Peter Horby, Annette Fox, Hien Tran Nguyen, Hang Khanh Le Nguyen, Phuong Mai Vu Hoang, Khanh Cong Nguyen, Menno D. de Jong, Rienk E. Jeeninga, H. Rogier van Doorn, Jeremy Farrar, and Heiman F.L. Wertheim
Author affiliations: National Institute of Hygiene and Epidemiology, Hanoi, Vietnam (M.Q.Le, H.T. Nguyen, H.K.L. Nguyen, P.M.V. Hoang, K.C. Nguyen); Wellcome Trust Major Overseas Programme, Hanoi and Ho Chi Minh City, Vietnam (P. Horby, A. Fox, M.D. de Jong, H.R. van Doorn, J. Farrar, H.F.L. Wertheim); Centre for Tropical Medicine, Oxford, United Kingdom (P. Horby, A. Fox, H.R. van Doorn, J. Farrar, H.F.L. Wertheim); Academic Medical Center, Amsterdam, the Netherlands (M.D. de Jong, R.E. Jeeninga)
Main Article
Figure
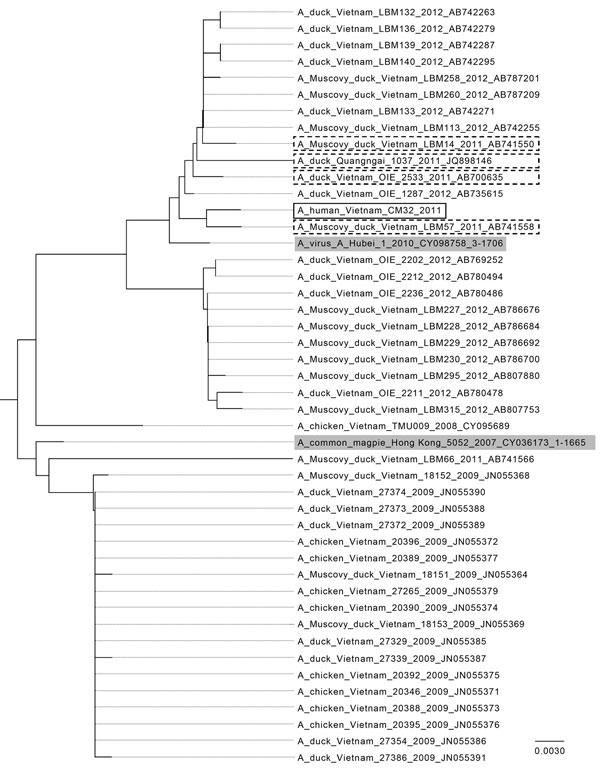
Figure. . Phylogentic analysis of avian influenza A(H5N1) virus clade 2.3.2.1 hemagglutinin DNA sequences from Vietnam compared with other isolates. Solid black box indicates isolate from the subclinical human case investigated in this study, A/CM32/2011; dashed boxes indicate sequences from Vietnam in 2011; gray shading indicates World Health Organization vaccine candidates A/common magpie/Hong Kong/5052/2007 and A/Hubei/1/2010 for clade 2.3.2.1. The sequences were downloaded from the Influenza Research Database (www.fludb.org), imported into MEGA 5.2 (www.megasoftware.net), and aligned by using MUSCLE (EMBL-EBI, Cambridgeshire, UK). The neighbor-joining tree was generated from the aligned sequences using standard settings. Scale bar indicates nucleotide substitutions per site.
Main Article
Page created: September 16, 2013
Page updated: September 16, 2013
Page reviewed: September 16, 2013
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
