Volume 19, Number 7—July 2013
Letter
Bartonella Species in Raccoons and Feral Cats, Georgia, USA
Cite This Article
Citation for Media
To the Editor: Bartonella spp. are vector-borne, facultative, intracellular bacteria that infect mammalian erythrocytes and endothelial cells and might cause chronic bacteremia and asymptomatic infections in reservoir hosts (1). There are currently 30–40 identified Bartonella species (2), and 14 of them are zoonotic; they have a wide variety of reservoirs, including rodents, carnivores, and ungulates (3). This study describes 2 Bartonella species in an urban population of raccoons and compares these findings to Bartonella infection in sympatric feral cats (Felis catus).
Raccoons (Procyon lotor) (n = 37) were live-trapped (Tomahawk Life Trap Company, Tomahawk, WI, USA) in spring and summer of 2012 on St. Simons Island, an urbanized coastal barrier island in Georgia in the southeastern United States (31°9′40″N, 81°23′13″W). The island is characterized by beach, salt marsh, forest, freshwater slough, and extensive residential developments. Raccoons were anesthetized with 20 mg/kg ketamine (Aveco Co., Fort Dodge, IA, USA) and 4 mg/kg xylazine (Mobay Corp., Shawnee, KS, USA), and blood was collected from the jugular vein into tubes containing EDTA. Feral cat blood samples (n = 37) from trap-neuter programs were collected by local veterinarians on St. Simons Island. Institutional Animal Care and Use Committee (A2011 03-042-Y2-A2) and Georgia Department of Natural Resources wildlife permits (29-WBH-12-100) were obtained before sampling.
DNA was extracted from blood by using a commercial DNA extraction kit (Quick-gDNA MiniPrep; Zymo Research Corp., Orange, CA, USA). Extracted DNA was used to amplify the 16S–23S rRNA intergenic spacer region of Bartonella spp. by nested PCR. For outer PCR, we used primers QHVE-1 (5′-TTCAGATGATGATCCCAAGC-3′) and QHVE-3 (5′-AACATGTCTGAATATATCTTC-3′) (4,5). PCR was performed with an initial incubation for 2 min at 94°C; 35 cycles of denaturation at 94°C for 30 s, primer annealing at 52°C for 30 s, and elongation at 72°C for 60 s; and a final incubation at 72°C for 6 min.
Nested PCR was performed by using primers QHVE-12 (5′-CCG GAG GGC TTG TAG CTC AG-3′) and QHVE-14b (5′-CCT CACAAT TTC AAT AGA AC-3′) (4). Nested PCR conditions were identical to those for the outer PCR, except for the annealing temperature, which was 55°C. Positive amplicons were separated by electrophoresis on a 1.2% agarose gel and purified by using the QIAquick PCR Purification Kit (QIAGEN, Valencia, CA, USA).
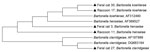
Purified DNA amplicons (400–600 bp) were sequenced by using an ABI automated sequencer (Applied Biosystems, Foster City, CA, USA). Intergenic spacer sequences from raccoon isolates were aligned with reported Bartonella species sequences in GenBank by using the ClustalW algorithm (6). A phylogenetic tree of the sequences was constructed by using neighbor-joining methods and maximum composite likelihood distances. Data were resampled 1,000 times to generate bootstrap values by using MEGA5 (7).
Of 74 samples analyzed (37 raccoon, 37 feral cat), 16 (43%) raccoon samples and 18 (48%) feral cat samples were positive for Bartonella spp. by PCR. Thirteen positive raccoon samples and 16 positive feral cat samples were sequenced. Twelve positive raccoon samples and 13 positive feral cat samples contained Bartonella henselae. B. koehlerae was amplified from 1 feral cat sample and 1 raccoon sample (99% sequence homology with a B. koehlerae sequence, GenBank accession no. AF312490). Two feral cat samples were identified as containing B. clarridgeiae and showed 98% and 100% sequence homology with a B. clarridgeiae sequence (GenBank accession no. AF167989) (Table; Figure, Appendix).
This study identified B. henselae and B. koehlerae in feral cat and raccoons and B. clarridgeiae in feral cats. Our results are useful because raccoons are potential reservoir hosts of zoonotic B. henselae and B. koehlerae, in addition to B. rochalimae, and there could be cross-species transmission of Bartonella spp. between feral cats and raccoons.
Among reservoir hosts for Bartonella species, rodents and cats have been the most extensively studied. Rodents harbor 11 Bartonella species (3). Cats are the principal reservoirs of B. clarridgeiae, which causes endocarditis in humans, and B. henselae, which causes cat-scratch disease. However, little is known about Bartonella spp. infections in raccoons; there is only 1 report of B. rochalimae in raccoons in California (8).
In this study, a relatively high proportion of raccoons were infected with B. henselae, implying that there is spillover of B. henselae from feral cats to raccoons or that raccoons are another active reservoir for B. henselae. B. clarridgeiae and B. koehlerae are also zoonotic; cats are primary reservoirs, and humans and dogs are accidental hosts (1). However, B. clarridgeiae was recently detected in rodent fleas in China (9) and B. koehlerae was isolated from feral pigs from the southeastern United States (10), suggesting that these pathogens also have multiple reservoir species.
Clarifying whether Bartonella infections in raccoons are caused by spillover from feral cats needs further study. Additional samples from raccoons and other species in urbanized and undeveloped habitats with different host species composition (e.g., cat-free environment) might enable further Bartonella spp. characterization in wildlife. We suspect urban raccoons and feral cats play a major role in Bartonella spp. transmission.
Acknowledgments
We thank Michael Yabsley, Barbara Shock, Matthew Stuckey, and Bruno Chomel for providing technical advice; and Mark Heth, Jan Rossiter and staff at the Island Animal Hospital, St. Simons Island, Georgia, for assistance with sample collection.
This study was supported by the St. Catherines Island Foundation, Inc., and Odum School of Ecology, University of Georgia, through the course taught by Vanessa Ezenwa and Andrew Park (ECOL/IDIS 8150L).
References
- Chomel BB, Boulouis HJ, Breitschwerdt EB, Kasten RW, Vayssier-Taussat M, Birtles RJ, Ecological fitness and strategies of adaptation of Bartonella species to their hosts and vectors. Vet Res. 2009;40:29. DOIPubMedGoogle Scholar
- Kosoy M, Hayman DT, Chan KS. Bartonella bacteria in nature: where does population variability end and a species start? Infect Genet Evol. 2012;12:894–904. DOIPubMedGoogle Scholar
- Kosoy MY. Ecological associations between bacteria of the genus Bartonella and mammals. Biol Bull. 2010;37:716–24. DOIGoogle Scholar
- Trataris AN, Rossouw J, Arntzen L, Karstaedt A, Frean J. Bartonella spp. in human and animal populations in Gauteng, South Africa, from 2007 to 2009. Onderstepoort J Vet Res. 2012;79:E1–8. DOIPubMedGoogle Scholar
- Roux V, Raoult D. The 16S–23S rRNA intergenic spacer region of Bartonella (Rochalimaea) species is longer than usually described in other bacteria. Gene. 1995;156:107–11. DOIPubMedGoogle Scholar
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. DOIPubMedGoogle Scholar
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. DOIPubMedGoogle Scholar
- Henn JB, Chomel BB, Boulouis HJ, Kasten RW, Murray WJ, Bar-Gal GK, Bartonella rochaelimae in raccoons, coyotes, and red foxes. Emerg Infect Dis. 2009;15:1984–7. DOIPubMedGoogle Scholar
- Li DM, Liu QY, Yu DZ, Zhang JZ, Gong ZD, Song XP. Phylogenetic analysis of Bartonella detected in rodent fleas in Yunnan, China. J Wildl Dis. 2007;43:609–17 .PubMedGoogle Scholar
- Beard AW, Maggi RG, Kennedy-Stoskopf S, Cherry NA, Sandfoss MR, DePerno CS, Bartonella spp. in feral pigs, southeastern United States. Emerg Infect Dis. 2011;17:893–5. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleRelated Links
Table of Contents – Volume 19, Number 7—July 2013
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Nicole Gottdenker, Department of Veterinary Pathology, College of Veterinary Medicine, University of Georgia, Athens, GA 30602, USA
Top