Volume 19, Number 8—August 2013
Letter
Novel Norovirus GII.4 Variant, Shanghai, China, 2012
Cite This Article
Citation for Media
To the Editor: Norovirus (NoV) has been identified as one of the major causal agents of nonbacterial, acute gastroenteritis in humans (1). The genetic diversity among NoVs is great, and human strains have been classified into 3 genogroups (GI, GII, and GIV). Despite this diversity, in recent years only a few strains, primarily those of genogroup II, genotype 4 (GII.4), have been responsible for most cases and outbreaks worldwide (1,2).
The pattern of epochal evolution of NoV is ongoing, and novel GII.4 variants emerge, which replace previously dominant strains and cause new pandemics. Surveillance systems worldwide showed an increase in NoV activity in late 2012 (3). Molecular data shared through NoroNet (www.rivm.nl/en/Topics/Topics/N/NoroNet) suggest that this increase is related to the emergence of a new GII.4 variant, termed Sydney_2012 (3). We found that this novel GII.4 variant also emerged in Shanghai, China, and caused increased levels of NoV activity during October–December 2012.
During July 2011–December 2012, fecal specimens from 748 outpatients (≥16 years of age) with acute gastroenteritis who visited 1 of the 2 sentinel hospitals in Shanghai were collected and stored at Shanghai Public Health Clinical Center at −70°C. Molecular detection of GI and GII NoV was performed by using conventional reverse transcription PCR as described (4). Full-length viral protein 1 and 639 bp of the 3′ RNA-dependent RNA polymerase gene of 4 randomly selected GII-positive strains were amplified (5–7). NoV genotypes were classified on the basis of a 280-bp region for GI and a 305-bp region for GII by using the Automated Genotyping Tool (www.rivm.nl/mpf/norovirus/typingtool).
A total of 77 patients showed positive results for GII NoV. An increase in GII NoV activity was observed during October–December in 2012; the detection rate was 46.08% (47 cases in 102 outpatients). The prevalence of GII NoV during the same period in 2011 was low; the detection rate was 6.90% (8 cases in 116 outpatients). Genotyping analysis of the strains detected in these 3 months in 2012 (39 strains were sequenced) showed that except for 1 GII.6 strain and 3 GII.4 2006b strains, the other 35 strains sequenced all belong to the new established cluster of GII.4, termed Sydney_2012. Retrospective analysis indicated that the novel GII.4 variant had already been detected in 2 outpatients during September 2011 in Shanghai.
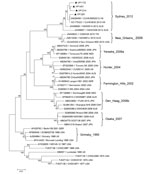
Phylogenetic analysis of full-length capsid nucleotide sequences for 4 strains randomly selected from the new cluster indicated a novel GII.4 pattern, and new strains clustering separately from previously identified GII.4 pandemic strains (Figure). On the basis of BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) searches, the most closely related NoVs (98%–100% nucleotide identity) were 4 GII.4 viruses recently detected in Australia and Hong Kong. The new GII.4 strains detected in Shanghai also clustered with these strains, a finding that was supported by bootstrap values >70% (Figure). The 3′ end of RNA-dependent RNA polymerase gene sequences also confirmed that the new GII.4 strains were recombinants, with a GII.e polymerase and GII.4 capsid (3).
Despite improved control measures to combat NOV, this highly infectious agent continues to cause a large number of epidemics of gastroenteritis globally (approximately every 2 years), and most epidemics have been associated with emergence of a novel GII.4 cluster (9). The new cluster reported in the present study was first detected in Australia in March, 2012, followed by detection in France, New Zealand, Japan, the United Kingdom, the United States, and Hong Kong, where increased levels of NoV activity in late 2012 compared with previous seasons were also observed (3). This novel GII.4 strain has also emerged in Shanghai, China, and caused increased levels of sporadic cases during October–and December 2012. This new variant has common ancestors, dominant NoV GII.4 variants Osaka_2007 and New Orleans_2009, but is phylogenetically distinct from them. Amino acid changes are present in major epitopes located in the P2 domain, a finding that is consistent with observations from previous epidemics (3).
Acknowledgment
This study was supported by grant (no. 2012ZX10004-211) from the Ministry of Health, People’s Republic of China for the National Major Science and Technology Project “Prevention and Treatment of AIDS, Viral Hepatitis, and Other Major Infectious Diseases.”
References
- Patel MM, Widdowson MA, Glass RI, Akazawa K, Vinje J, Parashar UD. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg Infect Dis. 2008;14:1224–31 . DOIPubMedGoogle Scholar
- Kroneman A, Verhoef L, Harris J, Vennema H, Duizer E, van Duynhoven Y, Analysis of integrated virological and epidemiological reports of norovirus outbreaks collected within the foodborne viruses in Europe network from 1 July 2001 to 30 June 2006. J Clin Microbiol. 2008;46:2959–65. DOIPubMedGoogle Scholar
- van Beek J, Ambert-Balay K, Botteldoorn N, Eden JS, Fonager J, Hewitt J, Indications for worldwide increased norovirus activity associated with emergence of a new variant of genotype II.4, late 2012. Euro Surveill. 2013;18:8–9 .PubMedGoogle Scholar
- Kojima S, Kageyama T, Fukushi S, Hoshino FB, Shinohara M, Uchida K, Genogroup-specific PCR primers for detection of Norwalk-like viruses. J Virol Methods. 2002;100:107–14. DOIPubMedGoogle Scholar
- Yuen LK, Catton MG, Cox BJ, Wright PJ, Marshall JA. Heminested multiplex reverse transcription-PCR for detection and differentiation of Norwalk-like virus genogroups 1 and 2 in fecal samples. J Clin Microbiol. 2001;39:2690–4. DOIPubMedGoogle Scholar
- Tu ET, Bull RA, Greening GE, Hewitt J, Lyon MJ, Marshall JA, Epidemics of gastroenteritis during 2006 were associated with the spread of norovirus GII.4 variants 2006a and 2006b. Clin Infect Dis. 2008;46:413–20. DOIPubMedGoogle Scholar
- Bull RA, Hansman GS, Clancy LE, Tanaka MM, Rawlinson WD, White PA. Norovirus recombination in ORF1/ORF2 overlap. Emerg Infect Dis. 2005;11:1079–85. DOIPubMedGoogle Scholar
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. DOIPubMedGoogle Scholar
- Siebenga JJ, Vennema H, Zheng DP, Vinje J, Lee BE, Pang XL, Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001–2007. J Infect Dis. 2009;200:802–12 . DOIPubMedGoogle Scholar
Figure
Cite This ArticleRelated Links
Table of Contents – Volume 19, Number 8—August 2013
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Jun Zhang, Department of Clinical Laboratory Medicine, Shanghai Public Health Clinical Center, Fudan University, 2901 Caolang Rd, Jinshan District, Shanghai, China
Top