Volume 20, Number 6—June 2014
Dispatch
Novel Phlebovirus with Zoonotic Potential Isolated from Ticks, Australia
Figure 2
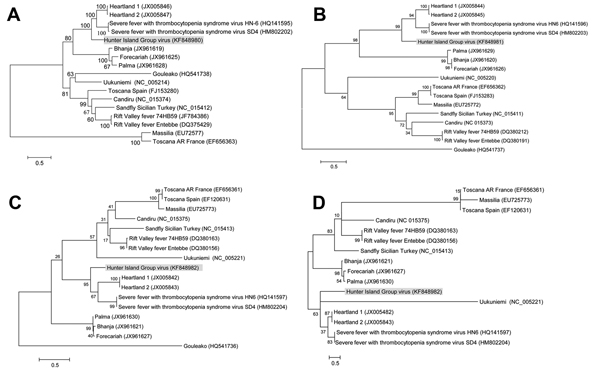
Figure 2. Phylogenetic trees of recently isolated bunyaviruses based on amino acid sequences of the polymerase protein (A) encoded by the large segment, the membrane glycoprotein polyprotein (B) encoded by the medium segment, and the nucleocapsid protein (C) and the nonstructural protein (D) encoded by the small segment of selected bunyavirusesMaximum-likelihood trees were constructed by using MEGA5 (http://www.megasoftware.net/) with bootstrapping at 1,000 replicatesGenBank accession numbers are within parentheses next to the virus namesScale bars indicate nucleotide substitutions per site.
1These authors contributed equally to this article.
Page created: May 16, 2014
Page updated: May 16, 2014
Page reviewed: May 16, 2014
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.