Volume 20, Number 7—July 2014
Dispatch
Widespread Rotavirus H in Commercially Raised Pigs, United States
Figure 2
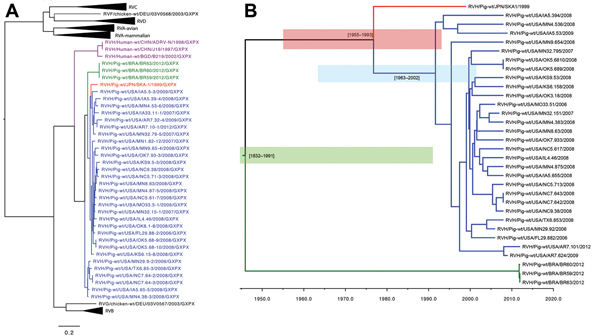
Figure 2. A) Nucleotide neighbor-joining phylogenetic tree of rotavirus (RV) A–D and F–H viral protein (VP) 6 sequencesBlue strains are from the United States; green strains are from Brazil; and the red strain is from JapanPurple strains are from humansScale bar indicates percentage of dissimilarity between sequencesB) Time-scaled phylogeny of swine RVH VP6 sequences using a Bayesian Markov chain Monte Carlo approachBlue shaded region indicates the time from the most recent common ancestor range (tMRCA) of the US strain; red shaded region indicates the US and Japan RVH tMCRA range; green shaded region indicates the tMRCA range for all swine RVH VP6 sequences.
Page created: June 17, 2014
Page updated: June 17, 2014
Page reviewed: June 17, 2014
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.