Volume 22, Number 10—October 2016
Letter
Recombinant Enterovirus A71 Subgenogroup C1 Strains, Germany, 2015
Figure
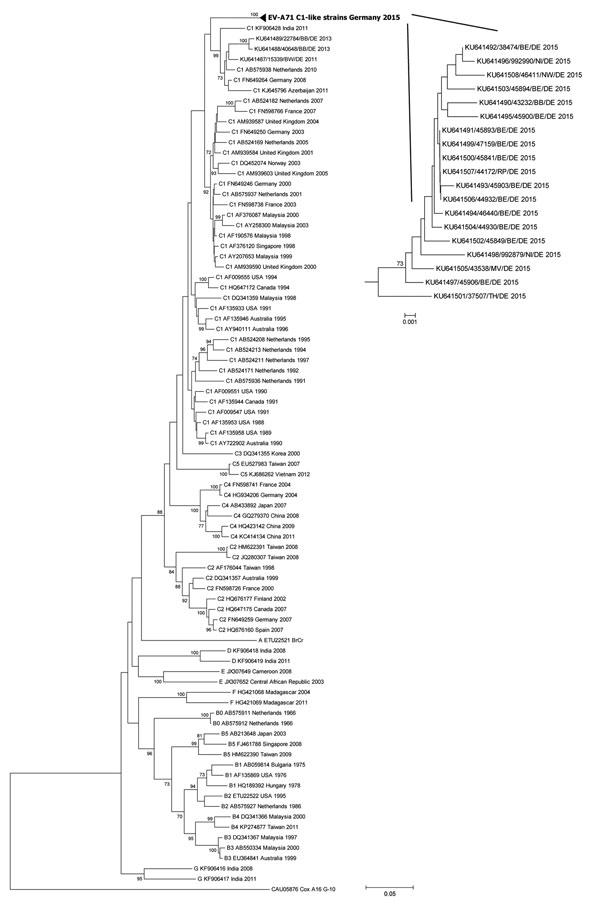
Figure. Phylogenetic tree based on complete viral protein 1 (VP1) nucleotide sequences of the strains identified within the German enterovirus surveillance (bold) and a representative set of enterovirus A71 strains available from GenBank (891 bases, corresponding to nucleotide positions 2439–3329 in the prototype BrCr ETU22521). The tree was constructed by using the neighbor-joining method (Kimura 2-parameter model) with 1,000 replicates through MEGA 6.06 (http://www.megasoftware.net/). Coxsackievirus A16 prototype (CAU05876) was used as the outgroup. Only bootstrap values >70 are shown. Genogroup and subgenogroup assignment, GenBank accession number, country and year of isolation are provided in the virus names. The enlarged subtree includes enterovirus A71 C1-like strains detected in Germany in 2015. Virus names contain strain number, abbreviation of federal state, country, and year of isolation. BE, Berlin; BB, Brandenburg; DE, Germany; MV, Mecklenburg Western Pomerania; NI, Lower Saxony; NW, Northrhine-Westphalia; RP, Rhineland Palatinate; TH, Thuringia. Scale bars indicate nucleotide substitution per site.
1Contributing members of the Laboratory Network for Enterovirus Diagnostics are listed at the end of this article.