Volume 22, Number 11—November 2016
Letter
Marseillevirus in the Pharynx of a Patient with Neurologic Disorders
Figure
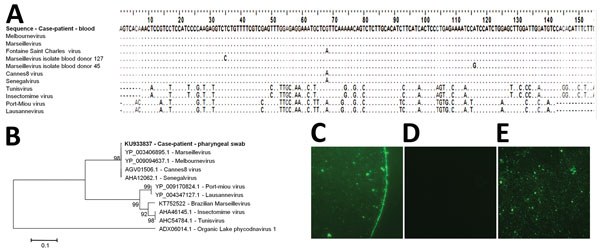
Figure. Marseillevirus sequences and serologic analysis for a 20-year-old man in Marseille, France, who initially sought treatment in November 2013 for a 2-day febrile gastroenteritis. A) Alignment of the sequence obtained in November 2014 from the blood of the case-patient with sequences from Marseillevirus and other related viruses. GenBank accession nos.: Marseillevirus, GU071086.1; Melbournevirus, KM275475.1; Fontaine Saint-Charles virus, KF582416.1; Senegalvirus, KF582412.1; Marseillevirus isolate blood donor 127, KF233993.1; Marseillevirus isolate blood donor 45, KF233992.1; Cannes 8 virus, KF261120.1; Tunisvirus, KF483846.1; Insectomime virus, KF527888.1; Port-Miou virus, KT428292.1; Lausannevirus, HQ113105.1. B) Phylogenetic reconstruction based on an amino acid alignment of the translated sequence obtained in November 2014 from a pharyngeal swab specimen from the case-patient (GenBank accession no. KU933837; indicated in bold) and homologous sequences from Marseilleviruses. Sequence from Organic Lake phycodnavirus 1 was used as an outgroup. The evolutionary history was inferred in MEGA6 software (http://www.megasoftware.net/) by the neighbor-joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. The evolutionary distances were computed by using the Kimura 2-parameter method. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Scale bar indicates nucleotide substitutions per site. C–E) Marseillevirus IgG detection by immunofluorescence in a serum sample from the case-patient. C) Serum sample from the case-patient at dilution 1:50; D) negative control (serum sample from a rabbit not exposed to Marseillevirus) at dilution 1:50; E) positive control (serum sample from a rabbit exposed to Marseillevirus) at dilution 1:50.