Volume 22, Number 11—November 2016
Research
Ambulatory Pediatric Surveillance of Hand, Foot and Mouth Disease as Signal of an Outbreak of Coxsackievirus A6 Infections, France, 2014–2015
Figure 4
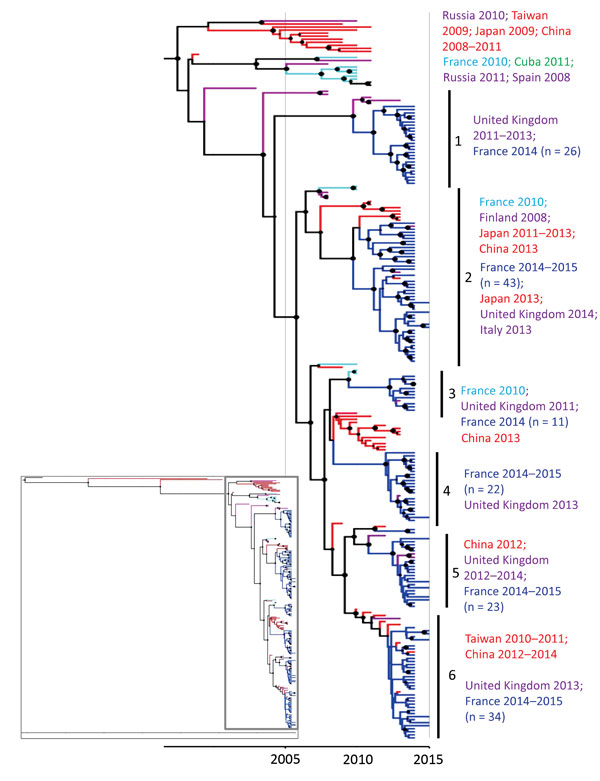
Figure 4. Phylogenetic tree based on partial viral protein (VP1) coding sequences of coxsackievirus (CV) A6, France, April 2014–March 2015. The maximum credibility tree is inferred with the partial VP1 sequence (369 nt, position 2,441–2,808 relative to the Gdula CV-A6 prototype strain). The phylogenetic relationships were inferred with a Bayesian method by using a relaxed molecular clock model. The tree was reconstructed using Figtree version 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree). For clarity, the sequence names are not included in the tree. Circle sizes are proportional to posterior probability. Each tip branch represents a sampled virus sequence. The continents/countries where the virus strains were sampled are indicated by different colors: Europe, purple; France 2010, light blue; France 2014–2015, dark blue; the Americas, green; Asia, red. The inset shows the complete tree, with the box indicating the portion enlarged for clarity.