Arenavirus Diversity and Phylogeography of Mastomys natalensis Rodents, Nigeria
Ayodeji Olayemi

, Adeoba Obadare, Akinlabi Oyeyiola, Joseph Igbokwe, Ayobami Fasogbon, Felix Igbahenah, Daniel Ortsega, Danny Asogun, Prince Umeh, Innocent Vakkai, Chukwuyem Abejegah, Meike Pahlman, Beate Becker-Ziaja, Stephan Günther, and Elisabeth Fichet-Calvet
Author affiliations: Obafemi Awolowo University, Ile-Ife, Nigeria (A. Olayemi, A. Obadare, A. Oyeyiola, J. Igbokwe); Ambrose Alli State University, Ekpoma, Nigeria (A. Fasogbon); Benue State University, Makurdi, Nigeria (F. Igbahenah, D. Ortsega); Irrua Specialist Teaching Hospital, Irrua, Nigeria (D. Asogun, C. Abejegah); Nigerian Montane Forest Project, Ngel-Nyaki, Nigeria (P. Umeh); Federal Medical Centre, Jalingo, Nigeria (I. Vakkai); Bernhard Nocht Institute for Tropical Medicine, Hamburg, Germany (M. Pahlman, B. Becker-Ziaja, S. Günther, E. Fichet-Calvet)
Main Article
Figure 2
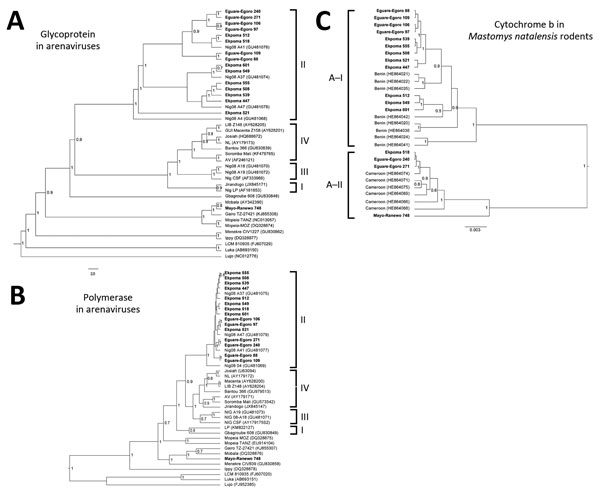
Figure 2. A) Phylogenetic analyses of glycoprotein precursor gene (GPC) of Old World arenaviruses and cytochrome b sequences of 16 arenavirus-positive Mastomys natalensis rodents captured in Nigeria during January 2011–March 2013 (boldface). The GPC tree (949 nt) was inferred by using the Bayesian Markov Chain Monte Carlo method, in a general time reversible plus gamma plus relaxed uncorrelated lognormal clock model. A random local clock was used for the cytochrome b tree. Bayesian posterior probabilities are shown at the node of the branches. The 4 lineages of the Lassa virus clade are indicated to the right of the GPC tree, and the 2 clades of M. natalensis rodents (A-I and A-II) are indicated on left of the cytochrome b tree. Scale bars indicate genetic distance. B) Phylogenetic analysis of the L (polymerase) gene in Old World arenaviruses, including the 16 new sequences found in M. natalensis rodents in Nigeria (boldface). The L tree (340 nt) was inferred by using the same method used for GPC analysis. GenBank numbers for reference isolate sequences are shown in parentheses.
Main Article
Page created: March 16, 2016
Page updated: March 16, 2016
Page reviewed: March 16, 2016
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
