Volume 22, Number 7—July 2016
Dispatch
Extended-Spectrum Cephalosporin-Resistant Salmonella enterica serovar Heidelberg Strains, the Netherlands1
Figure 2
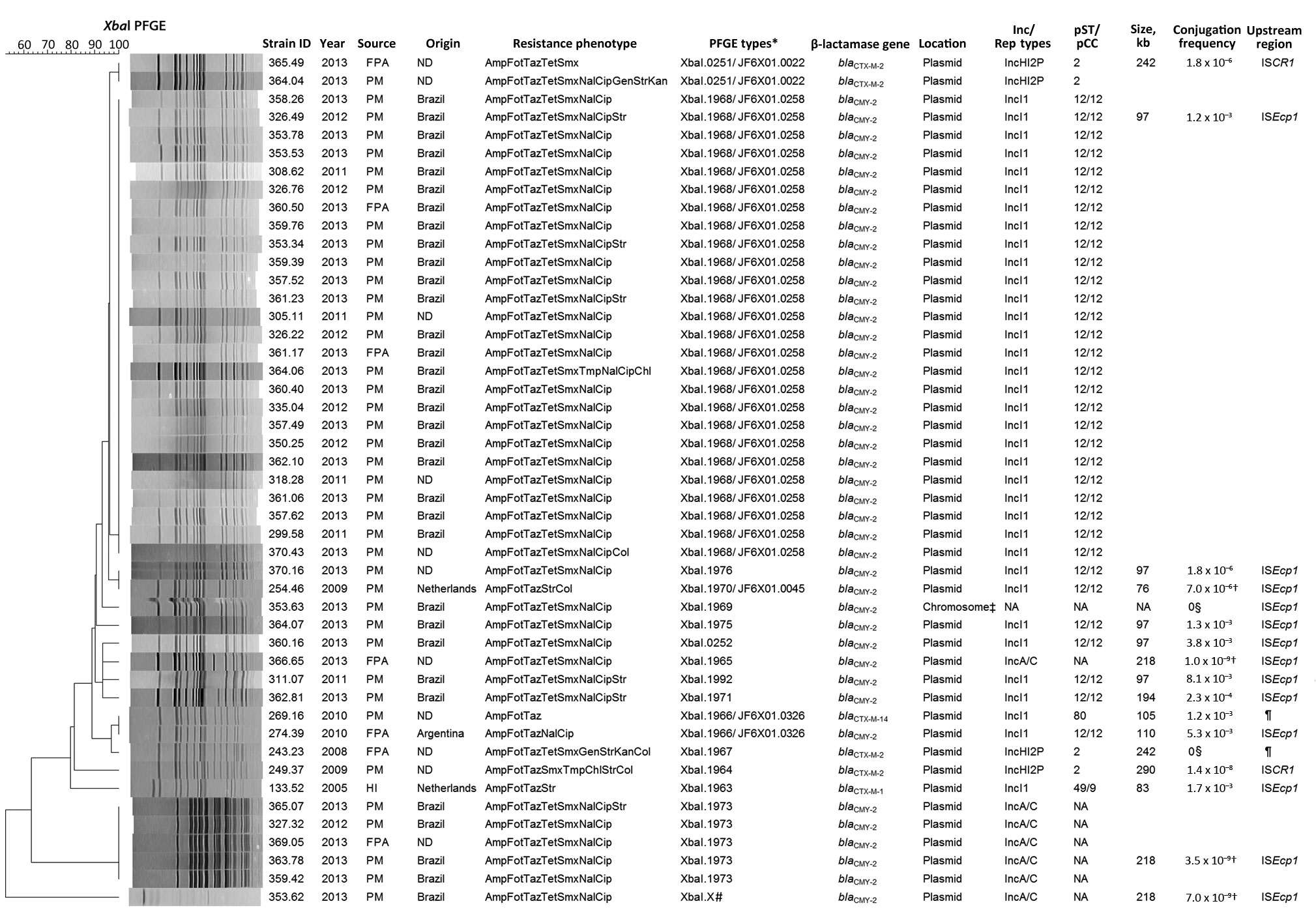
Figure 2. Characteristics of extended-spectrum cephalosporin-resistant Salmonella enterica serovar Heidelberg isolates, the Netherlands, 1999–2013. The dendrogram was generated by using BioNumerics version 6.6 (Applied Maths, Sint-Martens-Latem, Belgium) and indicates results of a cluster analysis on the basis of XbaI–pulsed-field gel electrophoresis (PFGE) fingerprinting. Similarity between the profiles was calculated with the Dice similarity coefficient and used 1% optimization and 1% band tolerance as position tolerance settings. The dendrogram was constructed with the UPGMA method based on the resulting similarity matrix. Amp, ampicillin; Cip, ciprofloxacin; Chl, chloramphenicol; Col, colistin; Fot, cefotaxime; FPA, food-producing animals; Gen, gentamicin; HI, human infection; Kan, kanamycin; Nal, nalidixic acid; ND, not determined (i.e., refers to isolates recovered in the Netherlands but with unknown origin of the sample); pCC, plasmid clonal complex; PM, poultry meat; pST, plasmid sequence type; Smx, sulfamethoxazole; Str, streptomycin; Taz, ceftazidime; Tet, tetracycline; Tmp, trimethoprim. *Pattern numbers assigned by The European Surveillance System molecular surveillance service of the European Centre for Disease Prevention and Control database and corresponding pattern numbers from the PulseNet database (http://www.cdc.gov/pulsenet/index.html). †Results refer to the conjugation frequencies during filter-mating experiments. ‡Chromosomal location confirmed by I-CeuI PFGE of total bacterial DNA, followed by Southern blot hybridization. §No transconjugants were obtained after liquid and filter-mating experiments, suggesting the presence of nonconjugative plasmids or conjugation frequencies below detection limits. ¶Insertion sequences ISEcp1, ISCR1, or IS26 were not found upstream of the extended-spectrum β-lactamase genes for these PFGE types. #This PFGE fingerprint was not submitted to The European Surveillance System molecular surveillance service of the European Centre for Disease Prevention and Control database for name assignment.
1Preliminary results from this study were presented at the 12th Beta-Lactamase Meeting, June 28–July 1, 2014, Gran Canaria, Spain.