Volume 22, Number 7—July 2016
Letter
Naturally Circulating Hepatitis A Virus in Olive Baboons, Uganda
Figure
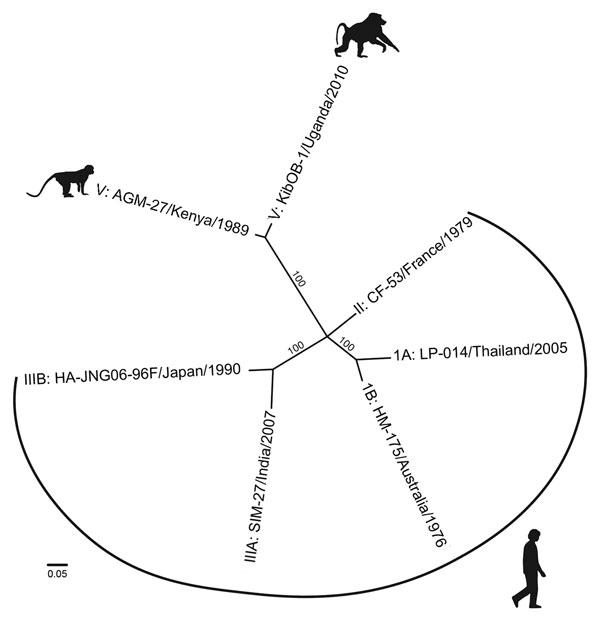
Figure. Whole-genome phylogenetic reconstruction of representative HAVs. HAVs are grouped into 6 genotypes based on 168 bp of the C-terminal extension of the viral protein 1 gene. Baboon HAV detected in Kibale National Park, Uganda, in 2010 and 2014 (GenBank accession number KT819575) clusters with AGM-27 (3), previously the sole member of genotype V. jModeltest 2 (http://jmodeltest.org) was used to find the best-fit evolutionary model for the data, after which the maximum-likelihood tree was estimated using the heuristic search method in PAUP* (http://paup.csit.fsu.edu), with starting trees obtained by neighbor-joining, random stepwise addition, and branch swapping by tree-bisection reconnection and starting branch lengths obtained using Rogers-Swofford approximation. Bootstrap values were derived from 1,000 replicates of the heuristic search; only values >50% are shown. GenBank accession nos.: IA, EF207320; IB, M14707; II, AY644676; IIIA, FJ227135; IIIB, AB258387; V, D00924). HAV, hepatitis A virus. Scale bar indicates substitutions per site.
References
- Dienstag JL, Davenport FM, McCollum RW, Hennessy AV, Klatskin G, Purcell RH. Nonhuman primate–associated viral hepatitis type A. Serologic evidence of hepatitis A virus infection. JAMA. 1976;236:462–4. DOIPubMedGoogle Scholar
- Robertson BH, Jansen RW, Khanna B, Totsuka A, Nainan OV, Siegl G, Genetic relatedness of hepatitis A virus strains recovered from different geographical regions. J Gen Virol. 1992;73:1365–77. DOIPubMedGoogle Scholar
- Tsarev SA, Emerson SU, Balayan MS, Ticehurst J, Purcell RH. Simian hepatitis A virus (HAV) strain AGM-27: comparison of genome structure and growth in cell culture with other HAV strains. J Gen Virol. 1991;72:1677–83. DOIPubMedGoogle Scholar
- Lauck M, Hyeroba D, Tumukunde A, Weny G, Lank SM, Chapman CA, Novel, divergent simian hemorrhagic fever viruses in a wild Ugandan red colobus monkey discovered using direct pyrosequencing. PLoS One. 2011;6:e19056. DOIPubMedGoogle Scholar
- Johnson CA, Swedell L, Rothman JM. Feeding ecology of olive baboons (Papio anubis) in Kibale National Park, Uganda: preliminary results on diet and food selection. Afr J Ecol. 2012;50:367–70.
- Emerson SU, Tsarev SA, Govindarajan S, Shapiro M, Purcell RH. A simian strain of hepatitis A virus, AGM-27, functions as an attenuated vaccine for chimpanzees. J Infect Dis. 1996;173:592–7. DOIPubMedGoogle Scholar
- Drexler JF, Corman VM, Lukashev AN, Brand JMA, van den Gmyl AP, Brünink S, Evolutionary origins of hepatitis A virus in small mammals. Proc Natl Acad Sci U S A. 2015;112:15190–5. DOIPubMedGoogle Scholar
- Drewe JA, O’Riain MJ, Beamish E, Currie H, Parsons S. Survey of infections transmissible between baboons and humans, Cape Town, South Africa. Emerg Infect Dis. 2012;18:298–301. DOIPubMedGoogle Scholar
- Goldberg TL, Gillespie TR, Rwego IB, Estoff EL, Chapman CA. Forest fragmentation as cause of bacterial transmission among nonhuman primates, humans, and livestock, Uganda. Emerg Infect Dis. 2008;14:1375–82. DOIPubMedGoogle Scholar
- Naughton-Treves L. Predicting patterns of crop damage by wildlife around Kibale National Park, Uganda. Conserv Biol. 1998;12:156–68 .DOIGoogle Scholar