Volume 22, Number 8—August 2016
Dispatch
Importation of Hybrid Human-Associated Trypanosoma cruzi Strains of Southern South American Origin, Colombia
Figure 2
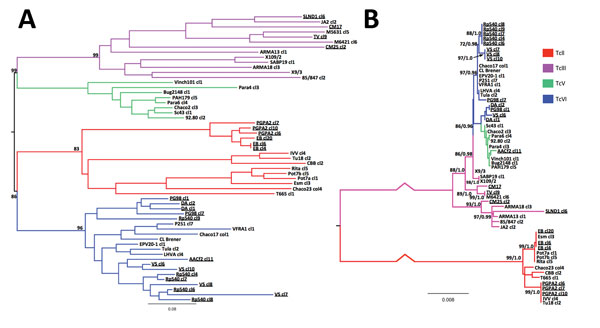
Figure 2. Phylogenetic trees showing relationships between Trypanosoma cruzi hybrids from Colombia and reference T. cruzi strains from across South America. A) Unrooted neighbor-joining tree based on pairwise distances between microsatellite loci. B) Maximum-likelihood tree from concatenated maxicircle sequences. Pairwise distance–based bootstrap values were calculated as the mean across 1,000 random diploid resamplings of the dataset; those >70% are shown for relevant nodes. A maximum-likelihood topology was constructed from concatenated maxicircle sequences for all clones. The most appropriate nucleotide substitution model was the general time reversible plus gamma distribution (9 substitution rate categories) based on the Akaike information criterion. Statistical support for major clades is given as equivalent bootstraps and posterior probabilities from consensus maximum-likelihood (1,000 pseudo-replicates) and Bayesian trees (based on the Hasegawa-Kishino-Yano plus gamma distribution model), respectively. Note that strain AACf2 cl11 is phylogenetically incongruent between nuclear and mitochondrial topologies. Branch colors indicate isolate discrete typing unit. Labels for clones from Colombia are underlined. Scale bars indicate genetic distance (A) and nucleotide substitutions per site (B).