Volume 23, Number 3—March 2017
Research
New Mycobacterium tuberculosis Complex Sublineage, Brazzaville, Congo
Figure 1
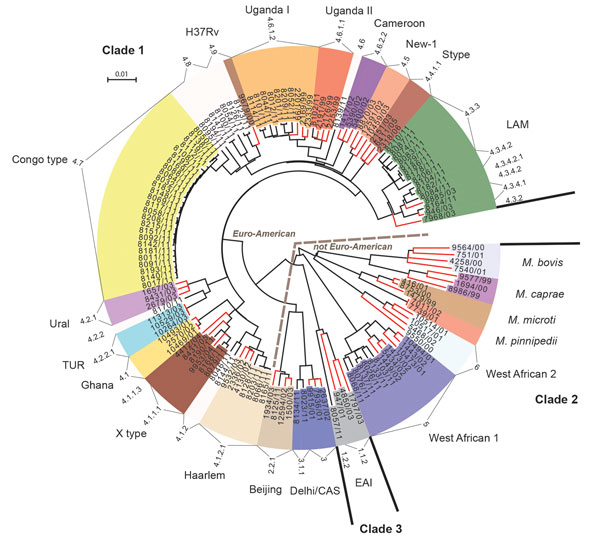
Figure 1. Maximum-likelihood tree of Mycobacterium tuberculosis complex isolates from Congo (black branch tips) and 65 reference strains (red branch tips). The tree was calculated by using the general time reversible substitution model with gamma distribution based on single-nucleotide polymorphisms identified by whole-genome sequencing. Models were tested and the tree generated by using MetaPiga software version 3.1 (18) and the maximum-likelihood ratio test. Midpoint rooting was performed. Distinct colors were chosen for the lineages identified; leaves with white background represent strains that initially were not assigned to particular lineages because of ambiguous typing patterns from mycobacterial interspersed repetitive unit, restriction fragment length polymorphism, or spoligo analysis (data not shown). The numerical code assigned to the respective lineages at the outer rim of the circular tree shows the Coll-nomenclature inferred from the whole-genome sequencing data. EAI, East African Indian; LAM, Latin American Mediterranean; TUR, Turkish. Scale bar indicates nucleotide substitutions per site.
References
- World Health Organization. Global tuberculosis report 2015. 20th ed. Geneva: The Organization; 2015.
- Comas I, Gagneux S. The past and future of tuberculosis research. PLoS Pathog. 2009;5:e1000600. DOIPubMedGoogle Scholar
- van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–86.PubMedGoogle Scholar
- Allix-Béguec C, Fauville-Dufaux M, Stoffels K, Ommeslag D, Walravens K, Saegerman C, et al. Importance of identifying Mycobacterium bovis as a causative agent of human tuberculosis. Eur Respir J. 2010;35:692–4. DOIPubMedGoogle Scholar
- Merker M, Blin C, Mona S, Duforet-Frebourg N, Lecher S, Willery E, et al. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat Genet. 2015;47:242–9. DOIPubMedGoogle Scholar
- Linguissi LS, Mayengue PI, Sidibé A, Vouvoungui JC, Missontsa M, Madzou-Laboum IK, et al. Prevalence of national treatment algorithm defined smear positive pulmonary tuberculosis in HIV positive patients in Brazzaville, Republic of Congo. BMC Res Notes. 2014;7:578. DOIPubMedGoogle Scholar
- Deutsches Institut für Normung. Medical microbiology. Diagnosis of tuberculosis. Part 3: detection of mycobacteria by culture methods. DIN 58943–3. Berlin; Beuth Verlag; 2011.
- Kent PT, Kubica GP. Public health mycobacteriology: a guide for the level III laboratory. Atlanta: Centers for Disease Control; 1985.
- Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–14.PubMedGoogle Scholar
- Gori A, Bandera A, Marchetti G, Degli Esposti A, Catozzi L, Nardi GP, et al. Spoligotyping and Mycobacterium tuberculosis. Emerg Infect Dis. 2005;11:1242–8. DOIPubMedGoogle Scholar
- Allix-Béguec C, Harmsen D, Weniger T, Supply P, Niemann S. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J Clin Microbiol. 2008;46:2692–9. DOIPubMedGoogle Scholar
- Weniger T, Krawczyk J, Supply P, Niemann S, Harmsen D. MIRU-VNTRplus: a Web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res. 2010;38(Web Server issue):W326–331.
- Blom J, Jakobi T, Doppmeier D, Jaenicke S, Kalinowski J, Stoye J, et al. Exact and complete short-read alignment to microbial genomes using Graphics Processing Unit programming. Bioinformatics. 2011;27:1351–8. DOIPubMedGoogle Scholar
- Roetzer A, Diel R, Kohl TA, Rückert C, Nübel U, Blom J, et al. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med. 2013;10:e1001387. DOIPubMedGoogle Scholar
- Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, Kremer K, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42:498–503. DOIPubMedGoogle Scholar
- Walker TM, Ip CL, Harrell RH, Evans JT, Kapatai G, Dedicoat MJ, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2013;13:137–46. DOIPubMedGoogle Scholar
- Coll F, McNerney R, Guerra-Assunção JA, Glynn JR, Perdigão J, Viveiros M, et al. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun. 2014;5:4812. DOIPubMedGoogle Scholar
- Helaers R, Milinkovitch MC. MetaPIGA v2.0: maximum likelihood large phylogeny estimation using the metapopulation genetic algorithm and other stochastic heuristics. BMC Bioinformatics. 2010;11:379. DOIPubMedGoogle Scholar
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–8. DOIPubMedGoogle Scholar
- Zhang H, Gao S, Lercher MJ, Hu S, Chen W-H. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 2012;40(W1):W569-72. DOIPubMedGoogle Scholar
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73. DOIPubMedGoogle Scholar
- Homolka S, Projahn M, Feuerriegel S, Ubben T, Diel R, Nübel U, et al. High resolution discrimination of clinical Mycobacterium tuberculosis complex strains based on single nucleotide polymorphisms. PLoS One. 2012;7:e39855. DOIPubMedGoogle Scholar
- Tessema B, Beer J, Merker M, Emmrich F, Sack U, Rodloff AC, et al. Molecular epidemiology and transmission dynamics of Mycobacterium tuberculosis in Northwest Ethiopia: new phylogenetic lineages found in Northwest Ethiopia. BMC Infect Dis. 2013;13:131. DOIPubMedGoogle Scholar
- Olaru ID, Rachow A, Lange C, Ntinginya NE, Reither K, Hoelscher M, et al. Ascertaining in vivo virulence of Mycobacterium tuberculosis lineages in patients in Mbeya, Tanzania. Int J Tuberc Lung Dis. 2015;19:70–3. DOIPubMedGoogle Scholar
- Niobe-Eyangoh SN, Kuaban C, Sorlin P, Cunin P, Thonnon J, Sola C, et al. Genetic biodiversity of Mycobacterium tuberculosis complex strains from patients with pulmonary tuberculosis in Cameroon. J Clin Microbiol. 2003;41:2547–53. DOIPubMedGoogle Scholar
- Koro Koro F, Kamdem Simo Y, Piam FF, Noeske J, Gutierrez C, Kuaban C, et al. Population dynamics of tuberculous Bacilli in Cameroon as assessed by spoligotyping. J Clin Microbiol. 2013;51:299–302. DOIPubMedGoogle Scholar
- Sidze LK, Tekwu EM, Kuaban C, Assam J-PA, Tedom J-C, Niemann S, et al. Estimates of genetic variability of Mycobacterium tuberculosis complex and its association with drug resistance in Cameroon. Adv Infect Dis. 2013;3:55. DOIGoogle Scholar
- Mbugi EV, Katale BZ, Streicher EM, Keyyu JD, Kendall SL, Dockrell HM, et al. Mapping of Mycobacterium tuberculosis complex genetic diversity profiles in Tanzania and other African countries. PLoS One. 2016;11:e0154571. DOIPubMedGoogle Scholar
- Hermans PW, Messadi F, Guebrexabher H, van Soolingen D, de Haas PE, Heersma H, et al. Analysis of the population structure of Mycobacterium tuberculosis in Ethiopia, Tunisia, and The Netherlands: usefulness of DNA typing for global tuberculosis epidemiology. J Infect Dis. 1995;171:1504–13. DOIPubMedGoogle Scholar
- Gagneux S, Long CD, Small PM, Van T, Schoolnik GK, Bohannan BJM. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science. 2006;312:1944–6. DOIPubMedGoogle Scholar
- Victor TC, de Haas PEW, Jordaan AM, van der Spuy GD, Richardson M, van Soolingen D, et al. Molecular characteristics and global spread of Mycobacterium tuberculosis with a western cape F11 genotype. J Clin Microbiol. 2004;42:769–72. DOIPubMedGoogle Scholar
- van Soolingen D, Qian L, de Haas PE, Douglas JT, Traore H, Portaels F, et al. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J Clin Microbiol. 1995;33:3234–8.PubMedGoogle Scholar
- Park YK, Bai GH, Kim SJ. Restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolated from countries in the western pacific region. J Clin Microbiol. 2000;38:191–7.PubMedGoogle Scholar
- Asiimwe BB, Koivula T, Källenius G, Huard RC, Ghebremichael S, Asiimwe J, et al. Mycobacterium tuberculosis Uganda genotype is the predominant cause of TB in Kampala, Uganda. Int J Tuberc Lung Dis. 2008;12:386–91.PubMedGoogle Scholar
- Feuerriegel S, Oberhauser B, George AG, Dafae F, Richter E, Rüsch-Gerdes S, et al. Sequence analysis for detection of first-line drug resistance in Mycobacterium tuberculosis strains from a high-incidence setting. BMC Microbiol. 2012;12:90. DOIPubMedGoogle Scholar
- Aubry A, Sougakoff W, Bodzongo P, Delcroix G, Armand S, Millot G, et al. First evaluation of drug-resistant Mycobacterium tuberculosis clinical isolates from Congo revealed misdetection of fluoroquinolone resistance by line probe assay due to a double substitution T80A-A90G in GyrA. PLoS One. 2014;9:e95083. DOIPubMedGoogle Scholar
- Sidze LK, Mouafo Tekwu E, Kuaban C, Assam Assam J-P, Tedom J-C, Eyangoh S, et al. Strong decrease in streptomycin-resistance and absence of XDR 12 years after the Reorganization of the National Tuberculosis Control Program in the Central Region of Cameroon. PLoS One. 2014;9:e98374. DOIPubMedGoogle Scholar
- Tekwu EM, Sidze LK, Assam J-P, Tedom J-C, Tchatchouang S, Makafe GG, et al. Sequence analysis for detection of drug resistance in Mycobacterium tuberculosis complex isolates from the Central Region of Cameroon. BMC Microbiol. 2014;14:113. DOIPubMedGoogle Scholar