Volume 23, Number 7—July 2017
Dispatch
Detection and Genetic Characterization of Adenovirus Type 14 Strain in Students with Influenza-Like Illness, New York, USA, 2014–2015
Figure 1
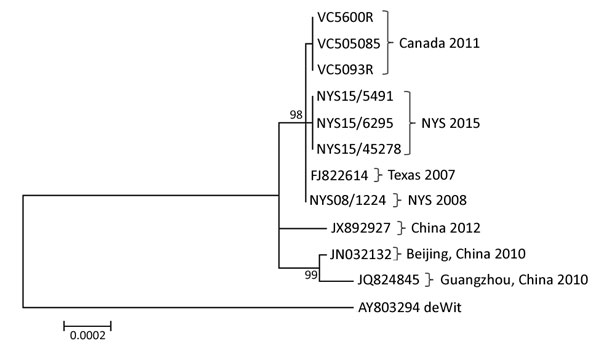
Figure 1. Phylogenetic tree of human adenoviruses constructed using 7 sequences obtained from college students with influenza-like illness, New York, USA, 2014–2015, and reference sequences of isolates from China (GenBank accession nos. JX892927, JN032132, and JQ824845); an isolate from Texas, USA (accession no. FJ822614); and the prototype strain, de Wit, from the Netherlands (accession no. AY803294). The tree was created by using the maximum-likelihood method based on the Kimura 2-parameter model with 500 bootstrap replicates from the whole-genome sequence of the displayed sequences. Pairwise distances were estimated by using the maximum composite–likelihood approach. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA6 (14). Scale bar indicates number of substitutions per site. NYS, New York State.
References
- Hage E, Gerd Liebert U, Bergs S, Ganzenmueller T, Heim A. Human mastadenovirus type 70: a novel, multiple recombinant species D mastadenovirus isolated from diarrhoeal faeces of a haematopoietic stem cell transplantation recipient. J Gen Virol. 2015;96:2734–42. DOIPubMedGoogle Scholar
- Harrach B, Benkő M, Both GW, Brown M, Davison AJ, Echavarría M, et al. Virus taxonomy: classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. 2011;2011:125–41.
- Kajon AE, Lu X, Erdman DD, Louie J, Schnurr D, George KS, et al. Molecular epidemiology and brief history of emerging adenovirus 14-associated respiratory disease in the United States. J Infect Dis. 2010;202:93–103. DOIPubMedGoogle Scholar
- Lewis PF, Schmidt MA, Lu X, Erdman DD, Campbell M, Thomas A, et al. A community-based outbreak of severe respiratory illness caused by human adenovirus serotype 14. J Infect Dis. 2009;199:1427–34. DOIPubMedGoogle Scholar
- Tate JE, Bunning ML, Lott L, Lu X, Su J, Metzgar D, et al. Outbreak of severe respiratory disease associated with emergent human adenovirus serotype 14 at a US air force training facility in 2007. J Infect Dis. 2009;199:1419–26. DOIPubMedGoogle Scholar
- Esposito DH, Gardner TJ, Schneider E, Stockman LJ, Tate JE, Panozzo CA, et al. Outbreak of pneumonia associated with emergent human adenovirus serotype 14—Southeast Alaska, 2008. J Infect Dis. 2010;202:214–22. DOIPubMedGoogle Scholar
- Mi Z, Butt AM, An X, Jiang T, Liu W, Qin C, et al. Genomic analysis of HAdV-B14 isolate from the outbreak of febrile respiratory infection in China. Genomics. 2013;102:448–55. DOIPubMedGoogle Scholar
- O’Flanagan D, O’Donnell J, Domegan L, Fitzpatrick F, Connell J, Coughlan S, et al. First reported cases of human adenovirus serotype 14p1 infection, Ireland, October 2009 to July 2010. Euro Surveill. 2011;16:19801.PubMedGoogle Scholar
- Parcell BJ, McIntyre PG, Yirrell DL, Fraser A, Quinn M, Templeton K, et al. Prison and community outbreak of severe respiratory infection due to adenovirus type 14p1 in Tayside, UK. J Public Health (Oxf). 2015;37:64–9. DOIPubMedGoogle Scholar
- Girouard G, Garceau R, Thibault L, Oussedik Y, Bastien N, Li Y. Adenovirus serotype 14 infection, New Brunswick, Canada, 2011. Emerg Infect Dis. 2013;19:119–22. DOIPubMedGoogle Scholar
- Okada M, Ogawa T, Kubonoya H, Yoshizumi H, Shinozaki K. Detection and sequence-based typing of human adenoviruses using sensitive universal primer sets for the hexon gene. Arch Virol. 2007;152:1–9. DOIPubMedGoogle Scholar
- Xu W, McDonough MC, Erdman DD. Species-specific identification of human adenoviruses by a multiplex PCR assay. J Clin Microbiol. 2000;38:4114–20.PubMedGoogle Scholar
- Kajon AEED. Assessment of genetic variability among subspecies B1 human adenoviruses for molecular epidemiology studies. In: Wold WSM, Tollefson AE, editors. Methods in molecular medicine, volume 130: adenovirus methods and protocols: adenoviruses, ad vectors, quantitation, and animal models. Vol. 1, 2nd ed. In: Wold W TA, editor. Methods of molecular medicine: adenovirus methods and protocols. Totowa (NJ): Humana Press; 2007. p. 335–55.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. DOIPubMedGoogle Scholar
- Mölsä M, Hemmilä H, Rönkkö E, Virkki M, Nikkari S, Ziegler T. Molecular characterization of adenoviruses among finnish military conscripts. J Med Virol. 2016;88:571–7. DOIPubMedGoogle Scholar