Volume 23, Number 7—July 2017
Research Letter
Novel Avulaviruses in Penguins, Antarctica
Abstract
We identified 3 novel and distinct avulaviruses from Gentoo penguins sampled in Antarctica. We isolated these viruses and sequenced their complete genomes; serologic assays demonstrated that the viruses do not have cross-reactivity between them. Our findings suggest that these 3 new viruses represent members of 3 novel avulavirus species.
Avian paramyxovirus (APMV) belongs to the genus Avulavirus, family Paramyxoviridae. There are 13 recognized Avulavirus species, each with 1 member, called avian paramyxovirus 1–13 (APMV-1–APMV-13) (1). A putative APMV-14 also has been recently described but not yet formally recognized (2).
In the past decade, APMV-10 through APMV-14 have been reported because of the intensification of surveillance of avian influenza A viruses (3–6). Most of the avulaviruses have been detected in wild birds associated with mild or no clinical disease; only Newcastle disease virus (a strain of APMV-1), APMV-2, and APMV-3 might cause substantial disease in poultry (7). Previous studies have described the presence of APMV-1, APMV-3, APMV-7, APMV-8, and other as-yet uncharacterized avulaviruses in Antarctic penguins (8). As a part of avian influenza surveillance expeditions in Antarctica during 2014–2016, we identified 3 novel avulaviruses in Gentoo penguins.
Cloacal, fecal, and serum samples were collected from Gentoo penguins (Pygoscelis papua) and Adélie penguins (P. adeliae), at 7 Antarctic locations (Technical Appendix Figure 1) during 2014–2016. Diagnostic tests, virus isolation, and serologic assays confirmed the identity of these paramyxoviruses (Technical Appendix).
We successfully isolated virus from 12 cloacal samples from Gentoo penguins on Kopaitic Island; these viruses showed positive hemagglutination titers ranging from 4 to 128 hemagglutination units. From these 12 isolates, only 5 were further confirmed by reverse transcription PCR and Sanger sequencing (9), suggesting the presence of new avulaviruses. All PCR-positive isolates were pooled and submitted for next-generation sequencing by using MiSeq 250 paired cycle run (Illumina, San Diego, CA, USA) (10).
By using next-generation sequencing, we obtained the genomic sequences of 3 novel avulaviruses that were named as follows: Antarctic penguin virus A (APVA), Antarctic penguin virus B (APVB), and Antarctic penguin virus C (APVC) (GenBank accession nos. KY452442–KY452444). Genome lengths of the 3 new avulaviruses ranged from 14,926 to 15,071 nt. The 6 genes for avulaviruses (coding for the nucleoprotein, phosphoprotein, matrix protein, fusion protein, hemagglutinin-neuraminidase protein, and RNA-dependent RNA polymerase protein) were identified in these virus genomes (Technical Appendix Figure 2, panel A). The sequence assembly was validated by coverage mapping (Technical Appendix Figure 2, panel B). The genomes described here are coding-complete; future experiments are needed to sequence the absolute terminus of the nontranslating region.
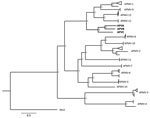
The 3 avulaviruses reported in this study showed 57%–60% genome-wide nucleotide identities to all other avulaviruses, as well as 32%–50% protein identities in the hemagglutinin-neuraminidase protein gene and 31%–48% in the fusion protein gene (Technical Appendix Figure 2, panel C). These new avulaviruses have 64%–67% genome-wide identity among each other. Accordingly, phylogenetic analyses (whether conducted by using genomes or specific genes) revealed that the new viruses form a monophyletic cluster with APMV-1, APMV-9, APMV-12, and APMV-13 (Figure; Technical Appendix Figure 3). Recently, a cutoff of <60% identity of nucleotide distance on whole genome has been proposed to differentiate avulaviruses (3); however, APMV-12 and APMV-13 and these 3 newly discovered viruses have higher identity. Thus, we suggest that this criterion requires further validation.
Phylogenetic analysis and pairwise comparison suggests that APVA, APVB, and APVC might each represent novel avulavirus species, which we recommend naming Avian avulavirus 15, 16, and 17, respectively (pending approval by the International Committee on Taxonomy of Viruses). We performed a hemagglutination inhibition assay by using APMV-1, APMV-2, APMV-3, APVA, and APVC antisera against isolates confirmed. No cross-reactivity was observed between APVA, APVB, and APVC. These viruses also did not show cross-reactivity against APMV-1, APMV2, and APMV-3 antisera. Antigenic results support the idea that novel viruses are 3 distinct species.
We observed cytopathic effects during infection of MDBK cells and Vero cells in all isolates evaluated. These effects were characterized by cell rounding and detachment of the monolayer, but syncytia were not evident (Technical Appendix Figure 4).
We also performed a hemagglutination inhibition assay by using APVA and APVC viruses. Three serum samples from Adélie penguins from Kopaitic Island reacted against APVC (titers 10–40), and 1 reacted against APVA (titer 40) (Technical Appendix Table). This result suggests that these novel avulaviruses can also infect Adélie penguins.
We report the successful virus isolation and whole-genome sequencing of avulaviruses in Antarctic penguin populations. Our analyses show that these viruses are genetically and antigenically divergent, indicating that Antarctic penguins harbor multiple avulaviruses. An important limitation is that the new viruses were not tested serologically against APMV-4 through APMV-13; however, genetic and antigenic differences between the new viruses support the idea that they are new species.
These data suggest that in Antarctica a much greater diversity of avulaviruses exists than previously recognized. Therefore, additional studies to evaluate the presence of these new viruses in other birds in Antarctica are needed to better understand the ecology and transmission of avulaviruses in this pristine environment.
Dr. Neira is assistant professor at the Animal Virology Unit, Facultad de Ciencias Veterinarias y Pecuarias, Universidad de Chile in Santiago. His research interests include understanding the diversity, transmission, and ecology of emerging and reemerging viruses in wildlife and livestock animals.
Acknowledgments
We thank the staff of Instituto Antártico Chileno for all their support during the expeditions to Antarctica, the Instituto de Salud Pública de Chile for biologic supplies, and The Chilean Navy and Antarctica XXI for assistance during field trips. We are grateful to Daniela Jiménez and Juan Mena for technical assistance. Sagar Goyal provided cell lines and APMV-1, APMV-2 and APMV-3 reference antisera.
This study is supported by the grants INACH RT46-16, RT12-13, CONICYT-PIA ANILLO ACT 1408, and FONDECYT 3150564, 11130305, 3150617, and 11160852.
References
- Afonso CL, Amarasinghe GK, Bányai K, Bào Y, Basler CF, Bavari S, et al. Taxonomy of the order Mononegavirales: update 2016. Arch Virol. 2016;161:2351–60. DOIPubMedGoogle Scholar
- Karamendin K, Kydyrmanov A, Seidalina A, Asanova S, Sayatov M, Kasymbekov E, et al. Complete genome sequence of a novel avian paramyxovirus (APMV-13) isolated from a wild bird in Kazakhstan. Genome Announc. 2016;4:e00167–16. DOIPubMedGoogle Scholar
- Thampaisarn R, Bui VN, Trinh DQ, Nagai M, Mizutani T, Omatsu T, et al. Characterization of avian paramyxovirus serotype 14, a novel serotype, isolated from a duck fecal sample in Japan. Virus Res. 2017;228:46–57. DOIPubMedGoogle Scholar
- Miller PJ, Afonso CL, Spackman E, Scott MA, Pedersen JC, Senne DA, et al. Evidence for a new avian paramyxovirus serotype 10 detected in rockhopper penguins from the Falkland Islands. J Virol. 2010;84:11496–504. DOIPubMedGoogle Scholar
- Briand F-X, Henry A, Massin P, Jestin V. Complete genome sequence of a novel avian paramyxovirus. J Virol. 2012;86:7710. DOIPubMedGoogle Scholar
- Goraichuk I, Sharma P, Stegniy B, Muzyka D, Pantin-Jackwood MJ, Gerilovych A, et al. Complete genome sequence of an avian paramyxovirus representative of putative new serotype 13. Genome Announc. 2016;4:e00729–16. DOIPubMedGoogle Scholar
- Alexander DJ. Newcastle disease and other avian paramyxoviruses. Rev Sci Tech. 2000;19:443–62. DOIPubMedGoogle Scholar
- Thomazelli LM, Araujo J, Oliveira DB, Sanfilippo L, Ferreira CS, Brentano L, et al. Newcastle disease virus in penguins from King George Island on the Antarctic region. Vet Microbiol. 2010;146:155–60. DOIPubMedGoogle Scholar
- van Boheemen S, Bestebroer TM, Verhagen JH, Osterhaus ADME, Pas SD, Herfst S, et al. A family-wide RT-PCR assay for detection of paramyxoviruses and application to a large-scale surveillance study. PLoS One. 2012;7:e34961. DOIPubMedGoogle Scholar
- Dill JA, Camus AC, Leary JH, Di Giallonardo F, Holmes EC, Ng TF. Distinct viral lineages from fish and amphibians reveal the 2 complex evolutionary history of hepadnaviruses. J Virol. 2016;90:7920–33. DOIPubMedGoogle Scholar
Figure
Cite This ArticleTable of Contents – Volume 23, Number 7—July 2017
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Victor Neira, Animal Virology Unit, Facultad de Ciencias Veterinarias y Pecuarias, Universidad de Chile, 11735 Santa Rosa, Santiago, Chile; ; Daniel Gonzalez Acuña, Zoology and Wildlife Laboratory, Facultad de Ciencias Veterinarias, Universidad de Concepción, 595 Vicente Méndez, Chillán, Chile
Top