Volume 23, Number 7—July 2017
Dispatch
Association of GII.P16-GII.2 Recombinant Norovirus Strain with Increased Norovirus Outbreaks, Guangdong, China, 2016
Figure 2
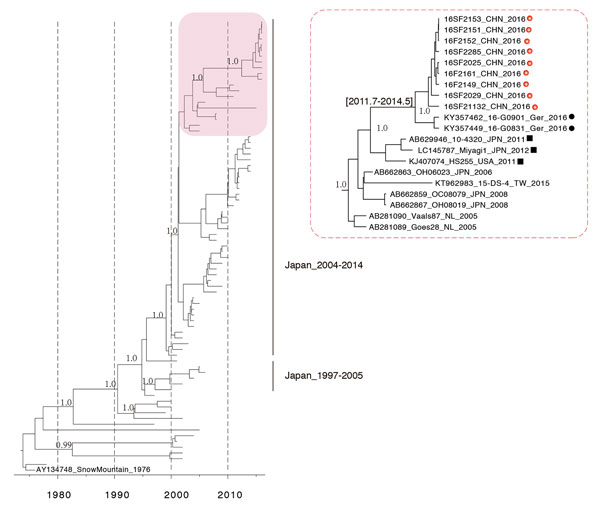
Figure 2. Molecular clock phylogeny of norovirus strain GII.2 VP1 gene sequences. The tree is a maximum clade credibility phylogeny with the GII.2 VP1 sequences, including the Guangdong, China, outbreak strains (red box, enlarged at right). Red dots indicate GII.2/Guangdong/2016 strains; black dots indicate outbreak strains from Germany, 2016; black squares indicate closely related GII.2 strains reported in previous years.
1These authors contributed equally to this article.
Page created: June 19, 2017
Page updated: June 19, 2017
Page reviewed: June 19, 2017
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.