Volume 23, Number 8—August 2017
Dispatch
Clonal Expansion of New Penicillin-Resistant Clade of Neisseria meningitidis Serogroup W Clonal Complex 11, Australia
Figure 1
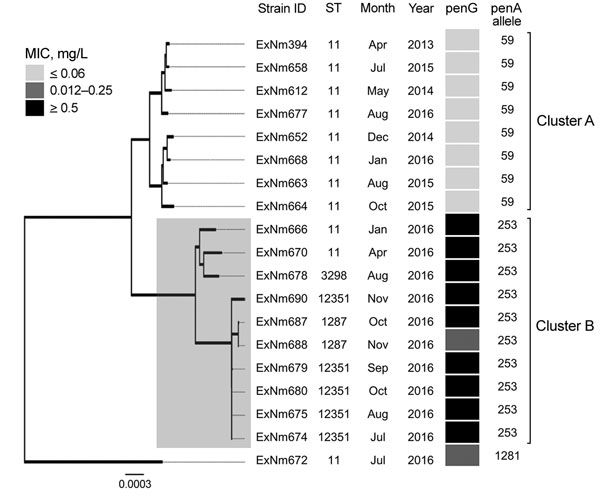
Figure 1. Neighbor-joining dendrogram (500 bootstrap values) for core genome sequences of clonal complex 11 Neisseria meningitidis strains with serogroup W capsules, Western Australia, Australia, January 2013–December 2016. The resistance phenotype for the penicillin G (PenG) gene for each isolate is provided using the following breakpoints: sensitive (MIC <0–0.06 mg/L), intermediate (0.12–0.25 mg/L) and resistant (>0.5 mg/L). Two clusters (A and B) were observed, which contain isolates that differ in penicillin resistance profile. Of 1,605 core-genome loci, a minimum of 244 are different between clusters A and B. The more recent cluster B appeared in early 2016 and contains penicillin-resistant isolates. Strain ExNm672 does not belong to either cluster. The dendrogram is drawn to scale, and sum of branch lengths between 2 strains indicates the proportion of nucleotide differences between those core genomes (≈1.5 Mb) within the pairwise alignment. Gray shaded box indicates isolates in cluster B. Scale bar indicates nucleotide substitutions per site. ID, identification; ST, sequence type.