Volume 24, Number 10—October 2018
Dispatch
Multilocus Sequence Typing of Mycoplasma pneumoniae, Japan, 2002–2016
Cite This Article
Citation for Media
Abstract
In Japan, Mycoplasma pneumoniae resistance to macrolides is high. To compare sequence types (STs) of susceptible and resistant isolates, we performed multilocus sequence typing for 417 isolates obtained in Japan during 2002–2016. The most prevalent ST overall was ST3, for macrolide-resistant was ST19, and for macrolide-susceptible were ST14 and ST7.
Macrolide-resistant Mycoplasma pneumoniae was first isolated in Japan in 2000 (1). Macrolide-resistant M. pneumoniae is highly prevalent in Asia and has been reported from several parts of the world (2–7). During a 2011–2012 outbreak in Japan, the resistance rate was as high as 90% (8). Molecular typing methods have been developed for M. pneumoniae and include multilocus variable-number tandem-repeat analysis (MLVA), P1 typing, and others (9). In recent years, multilocus sequence typing (MLST) involving molecular analysis of 7–8 housekeeping genes has been applied to various bacterial pathogens (10,11). In 2014, MLST for M. pneumoniae was devised by Brown et al. (12). Using this MLST method for M. pneumoniae, we compared sequence types (STs) of macrolide-susceptible and macrolide-resistant M. pneumoniae isolates from Japan and examined their evolutionary relationships. As is commonly performed for this pathogen, we also typed the P1 adhesin gene.
Using the random function of Excel 2010 (Microsoft, Redmond, CA, USA), we randomly selected 417 M. pneumoniae isolates (372 from children, 45 from adults) from 1,084 isolates obtained from patients throughout Japan who had had pneumonia or bronchitis during 2002–2016. Samples were not collected in 2014. The strains were isolated from clinical samples sent from 31 medical institutions in Japan to the Department of Infectious Diseases at Keio University School of Medicine in Tokyo. Of these strains, 232 (55.6%) were macrolide-susceptible M. pneumoniae and 185 (44.4%) were macrolide-resistant M. pneumoniae that had 23S rRNA point mutations. According to M. pneumoniae numbering, these mutations were A2063G (n = 163), A2063T (n = 10), A2064G (n = 10), A2063C (n = 1), or C2617A (n = 1). We used microdilution methods with pleuropneumonia-like organism broth (Difco, Detroit, MI, USA) to determine MICs for 6 antimicrobial agents against these strains of macrolide-susceptible and macrolide-resistant M. pneumoniae (Table 1).
For MLST analysis, we sequenced 8 housekeeping genes (ppa, pgm, gyrB, gmk, glyA, atpA, arcC, and adk) in each strain by using the 8 primer sets described in the MLST database (https://pubmlst.org/mpneumoniae/) (12). New alleles and STs were registered in the M. pneumoniae MLST database. To determine relationships between STs, we conducted clonal complex (CC) analysis by using eBURST version 3.1 (http://eburst.mlst.net/v3/mlst_datasets/). Frequency analysis was performed with the Fisher exact test.
Typing of the P1 adhesin gene in M. pneumoniae was performed as previously described (13). After PCR, the purified DNA products were treated with a restriction enzyme (HaeIII) for 1 hour. The standard stains M129 (ATCC 29342) and FH (ATCC 15531) were used as controls.
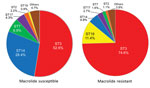
Among macrolide-susceptible M. pneumoniae isolates, the most prevalent STs were ST3 (52.6%), ST14 (28.4%), and ST7 (6.9%) (Figure 1). Among macrolide-resistant isolates, the most prevalent ST was also ST3 (74.6%), but the next most prevalent were ST19 (11.4%) and ST14 (4.9%). ST7 was more prevalent in macrolide-susceptible than in macrolide-resistant isolates (p = 0.008); ST19 was more prevalent in macrolide-resistant isolates (p<0.001). Although ST14 was prevalent among macrolide-susceptible M. pneumoniae, it also was represented among macrolide-resistant M. pneumoniae, associated with the 23S rRNA mutation A2064G. We did not identify any differences in ST frequencies between isolates from children and adults or between areas in Japan.
When we compared the relationship between year of strain isolation and ST (Table 2), ST3 accounted for most macrolide-susceptible M. pneumoniae, but ST14 also was present in every year; in other words, 2 STs were consistently common among macrolide-susceptible M. pneumoniae. In 2016, infection with macrolide-susceptible M. pneumoniae, especially ST7 and ST14, was epidemic. Among macrolide-resistant isolates, most were ST3. Although ST3 was identified more frequently in macrolide-susceptible isolates during 2002–2006, its identification in macrolide-resistant isolates has increased rapidly since then, and macrolide-resistant ST3 became more common in 2008. ST19 has been identified since the M. pneumoniae epidemic in 2006.
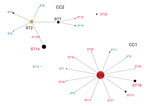
Data showing the relationship between CC and STs according to eBURST (Figure 2) include the 417 strains in this study and 62 strains registered to the MLST database from other countries. Of the 2 CC clusters, the center of CC1 was ST3, from which new STs including ST19 and ST17 arose with >1 alleles mutated. CC1 appears more frequently in macrolide-resistant M. pneumoniae (p<0.001). The center of CC2 was ST2, and almost all strains in Japan belonged to ST7 and ST14. ST22 did not belong to either CC1 or CC2, but 5 of 8 of its alleles were identical to those of CC1. In certain areas of Japan, ST22 has emerged in macrolide-resistant and macrolide-susceptible M. pneumoniae.
P1 typing analysis indicated that M. pneumoniae STs belonging to CC1 and ST22 were type 1, and STs belong to CC2 were type 2. ST14 and ST15 were type 2a, which is a subtype of P1 type 2.
Molecular analysis by MLST is conducted commonly for various bacterial pathogens worldwide but not for M. pneumoniae. We identified differences in STs between macrolide-susceptible and macrolide-resistant M. pneumoniae in Japan. Of the STs noted among isolates from 4 countries (the United Kingdom, the United States, China, and France), ST3 and ST14 were identified often in Japan, but other STs were not identified in Japan (Figure 2). During 1977–2011, the most prevalent M. pneumoniae STs in England and Wales were ST2, ST3, ST4, and ST11 (14). In this respect, STs reported from Japan show patterns quite different from those from other countries. The most prevalent STs in Japan were ST3, ST14, ST19, and ST7. In particular, ST3 of macrolide-susceptible M. pneumoniae was the most prevalent during 2002–2006, the early period of our surveillance, but ST3 of macrolide-resistant M. pneumoniae, with a point mutation of A2063G in the 23S rRNA gene, increased rapidly since an outbreak of M. pneumoniae infection in 2006, and macrolide-resistant M. pneumoniae became most common after 2008. This change toward resistance occurred in parallel with increased use of macrolides in children. ST14 and ST7 were more prevalent among macrolide-susceptible than macrolide-resistant M. pneumoniae; ST19 was prevalent among macrolide-resistant M. pneumoniae.
We detected 2 types of the P1 adhesin gene, types 1 and 2 (which includes 2a), and analyzed them in terms of STs. Recent studies have identified various subtypes through comprehensive genomic comparisons (15). Similar to the results of MLST, P1 gene types have shown diversification. We did not conduct MLVA in this study, but a previous study found CC1 and CC2 to differ in MLVA type: CC1 contained MLVA type 4572, and CC2 contained MLVA types 3662 and 3562 (12).
In summary, macrolide-resistant M. pneumoniae are emerging and increasing. Surveillance of macrolide-resistant M. pneumoniae based on molecular epidemiology, including STs and P1 adhesion gene identifications, are necessary to clarify worldwide trends and address public health concerns.
Ms. Ando is a graduate student in the Department of Infectious Diseases, Keio University School of Medicine. Dr. Morozumi is an assistant professor at Keio University School of Medicine; her research interests include molecular epidemiology as well as severe infections caused by Streptococcus agalactiae and M. pneumoniae.
Acknowledgment
We thank the pediatricians who actively participated in the Acute Respiratory Diseases Study Group. We are also grateful to respiratory physicians Tadashi Ishida, Hiroyuki Yoshimine, and Shigeo Hanada.
References
- Okazaki N, Narita M, Yamada S, Izumikawa K, Umetsu M, Kenri T, et al. Characteristics of macrolide-resistant Mycoplasma pneumoniae strains isolated from patients and induced with erythromycin in vitro. Microbiol Immunol. 2001;45:617–20. DOIPubMedGoogle Scholar
- Averbuch D, Hidalgo-Grass C, Moses AE, Engelhard D, Nir-Paz R. Macrolide resistance in Mycoplasma pneumoniae, Israel, 2010. Emerg Infect Dis. 2011;17:1079–82. DOIPubMedGoogle Scholar
- Eshaghi A, Memari N, Tang P, Olsha R, Farrell DJ, Low DE, et al. Macrolide-resistant Mycoplasma pneumoniae in humans, Ontario, Canada, 2010-2011. Emerg Infect Dis. 2013;19:19. DOIPubMedGoogle Scholar
- Li X, Atkinson TP, Hagood J, Makris C, Duffy LB, Waites KB. Emerging macrolide resistance in Mycoplasma pneumoniae in children: detection and characterization of resistant isolates. Pediatr Infect Dis J. 2009;28:693–6. DOIPubMedGoogle Scholar
- Morozumi M, Iwata S, Hasegawa K, Chiba N, Takayanagi R, Matsubara K, et al.; Acute Respiratory Diseases Study Group. Increased macrolide resistance of Mycoplasma pneumoniae in pediatric patients with community-acquired pneumonia. Antimicrob Agents Chemother. 2008;52:348–50. DOIPubMedGoogle Scholar
- Pereyre S, Charron A, Renaudin H, Bébéar C, Bébéar CM. First report of macrolide-resistant strains and description of a novel nucleotide sequence variation in the P1 adhesin gene in Mycoplasma pneumoniae clinical strains isolated in France over 12 years. J Clin Microbiol. 2007;45:3534–9. DOIPubMedGoogle Scholar
- Zhao F, Liu G, Wu J, Cao B, Tao X, He L, et al. Surveillance of macrolide-resistant Mycoplasma pneumoniae in Beijing, China, from 2008 to 2012. Antimicrob Agents Chemother. 2013;57:1521–3. DOIPubMedGoogle Scholar
- Okada T, Morozumi M, Tajima T, Hasegawa M, Sakata H, Ohnari S, et al. Rapid effectiveness of minocycline or doxycycline against macrolide-resistant Mycoplasma pneumoniae infection in a 2011 outbreak among Japanese children. Clin Infect Dis. 2012;55:1642–9. DOIPubMedGoogle Scholar
- Brown RJ, Spiller BO, Chalker VJ. Molecular typing of Mycoplasma pneumoniae: where do we stand? Future Microbiol. 2015;10:1793–5. DOIPubMedGoogle Scholar
- Enright MC, Spratt BG. Multilocus sequence typing. Trends Microbiol. 1999;7:482–7. DOIPubMedGoogle Scholar
- Skaare D, Anthonisen IL, Caugant DA, Jenkins A, Steinbakk M, Strand L, et al. Multilocus sequence typing and ftsI sequencing: a powerful tool for surveillance of penicillin-binding protein 3-mediated beta-lactam resistance in nontypeable Haemophilus influenzae. BMC Microbiol. 2014;20;14:131.
- Brown RJ, Holden MTG, Spiller OB, Chalker VJ. Development of a multilocus sequence typing scheme for molecular typing of Mycoplasma pneumoniae. J Clin Microbiol. 2015;53:3195–203. DOIPubMedGoogle Scholar
- Kenri T, Taniguchi R, Sasaki Y, Okazaki N, Narita M, Izumikawa K, et al. Identification of a new variable sequence in the P1 cytadhesin gene of Mycoplasma pneumoniae: evidence for the generation of antigenic variation by DNA recombination between repetitive sequences. Infect Immun. 1999;67:4557–62.https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=10456900&dopt=Abstract
- Brown RJ, Nguipdop-Djomo P, Zhao H, Stanford E, Spiller OB, Chalker VJ. Mycoplasma pneumoniae epidemiology in England and Wales: a national perspective. Front Microbiol. 2016;7:157. DOIPubMedGoogle Scholar
- Diaz MH, Desai HP, Morrison SS, Benitez AJ, Wolff BJ, Caravas J, et al. Comprehensive bioinformatics analysis of Mycoplasma pneumoniae genomes to investigate underlying population structure and type-specific determinants. PLoS One. 2017;14:12.
Figures
Tables
Cite This Article1These authors contributed equally to this article.
Table of Contents – Volume 24, Number 10—October 2018
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Satoshi Iwata, Keio University School of Medicine, Department of Infectious Diseases, 35 Shinanomachi, Shinjuku-ku, Tokyo 160-8582, Japan; and National Cancer Center Hospital, Department of Infectious Diseases, 5-1-1 Tsukiji, Chuo-ku, Tokyo 104-0045, Japan
Top