Volume 24, Number 11—November 2018
Research Letter
Burkholderia lata Infections from Intrinsically Contaminated Chlorhexidine Mouthwash, Australia, 2016
Figure
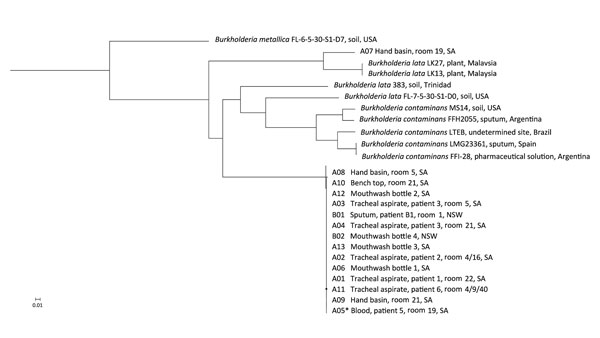
Figure. Phylogenetic analysis of isolates implicated in an outbreak Burkholderia lata infection from intrinsically contaminated chlorhexidine mouthwash, Australia, 2016. The maximum-likelihood tree is constructed from core genome single-nucleotide polymorphism alignments (N = 512,480) of the outbreak genomes, bootstrapped 1,000 times, and archival genomes from B. cepacia complex group K, relative to the reference genome B. lata A05 (identified by an asterisk). B. metallica was included as a comparator.
Page created: October 16, 2018
Page updated: October 16, 2018
Page reviewed: October 16, 2018
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.